Bacteria from the endosphere and rhizosphere of Quercus spp. use mainly cell wall-associated enzymes to decompose organic matter
- PMID: 30908541
- PMCID: PMC6433265
- DOI: 10.1371/journal.pone.0214422
Bacteria from the endosphere and rhizosphere of Quercus spp. use mainly cell wall-associated enzymes to decompose organic matter
Abstract
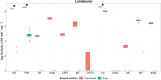
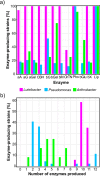
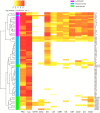
Due to the ability of soil bacteria to solubilize minerals, fix N2 and mobilize nutrients entrapped in the organic matter, their role in nutrient turnover and plant fitness is of high relevance in forest ecosystems. Although several authors have already studied the organic matter decomposing enzymes produced by soil and plant root-interacting bacteria, most of the works did not account for the activity of cell wall-attached enzymes. Therefore, the enzyme deployment strategy of three bacterial collections (genera Luteibacter, Pseudomonas and Arthrobacter) associated with Quercus spp. roots was investigated by exploring both cell-bound and freely-released hydrolytic enzymes. We also studied the potential of these bacterial collections to produce enzymes involved in the transformation of plant and fungal biomass. Remarkably, the cell-associated enzymes accounted for the vast majority of the total activity detected among Luteibacter strains, suggesting that they could have developed a strategy to maintain the decomposition products in their vicinity, and therefore to reduce the diffusional losses of the products. The spectrum of the enzymes synthesized and the titres of activity were diverse among the three bacterial genera. While cellulolytic and hemicellulolytic enzymes were rather common among Luteibacter and Pseudomonas strains and less detected in Arthrobacter collection, the activity of lipase was widespread among all the tested strains. Our results indicate that a large fraction of the extracellular enzymatic activity is due to cell wall-attached enzymes for some bacteria, and that Quercus spp. root bacteria could contribute at different levels to carbon (C), phosphorus (P) and nitrogen (N) cycles.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

