Diversity of transducer-like proteins (Tlps) in Campylobacter
- PMID: 30908544
- PMCID: PMC6433261
- DOI: 10.1371/journal.pone.0214228
Diversity of transducer-like proteins (Tlps) in Campylobacter
Abstract
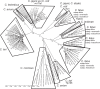
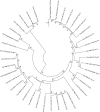
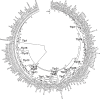
Campylobacter transducer-like proteins (Tlps), also known as methyl-accepting chemotaxis proteins (MCPs), are associated with virulence as well as niche and host adaptation. While functional attributes of these proteins are being elucidated, little has been published regarding their sequence diversity or chromosomal locations and context, although they appear to define invertible regions within Campylobacter jejuni genomes. Genome assemblies for several species of Campylobacter were obtained from the publicly available NCBI repositories. Genomes from all isolates were obtained from GenBank and assessed for Tlp content, while data from isolates with complete, finished genomes were used to determine the identity of Tlps as well as the gene content of putative invertible elements (IEs) in C. jejuni (Cj) and C. coli (Cc). Tlps from several Campylobacter species were organized into a nomenclature system and novel Tlps were defined and named for Cj and Cc. The content of Tlps appears to be species-specific, though diverse within species. Cj and Cc carried overlapping, related Tlp content, as did the three C. fetus subspecies. Tlp1 was detected in 88% of Cj isolates and approximately 43% of Cc, and was found in a different conserved chromosomal location and genetic context in each species. Tlp1 and Tlp 3 predominated in genomes from Cj whereas other Tlps were detected less frequently. Tlp13 and Tlp20 predominated in genomes from Cc while some Cj/Cc Tlps were not detected at all. Tlps 2-4 and 11-20 were less frequently detected and many showed sequence heterogeneity that could affect substrate binding, signal transduction, or both. Tlps other than Tlp1, 7, and 10 had substantial sequence identity in the C-terminal half of the protein, creating chromosomal repeats potentially capable of mediating the inversion of large chromosomal DNA. Cj and Cc Tlps were both found in association with only 14 different genes, indicating a limited genomic context. In Cj these Tlps defined IEs that were for the most part found at a single chromosomal location and comprised of a conserved set of genes. Cc IEs were situated at very different chromosomal locations, had different structures than Cj IEs, and were occasionally incomplete, therefore not capable of inversion. Tlps may have a role in Campylobacter genome structure and dynamics as well as acting as chemoreceptors mediating chemotactic responses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Casey E, Fitzgerald E, Lucey B. Towards understanding clinical campylobacter infection and its transmission: time for a different approach? Brit J Biomed Sci. 2017;74: 53–64. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

