Genetic Advances in Chronic Obstructive Pulmonary Disease. Insights from COPDGene
- PMID: 30908940
- PMCID: PMC6775891
- DOI: 10.1164/rccm.201808-1455SO
Genetic Advances in Chronic Obstructive Pulmonary Disease. Insights from COPDGene
Abstract
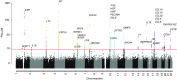
Chronic obstructive pulmonary disease (COPD) is a common and progressive disease that is influenced by both genetic and environmental factors. For many years, knowledge of the genetic basis of COPD was limited to Mendelian syndromes, such as alpha-1 antitrypsin deficiency and cutis laxa, caused by rare genetic variants. Over the past decade, the proliferation of genome-wide association studies, the accessibility of whole-genome sequencing, and the development of novel methods for analyzing genetic variation data have led to a substantial increase in the understanding of genetic variants that play a role in COPD susceptibility and COPD-related phenotypes. COPDGene (Genetic Epidemiology of COPD), a multicenter, longitudinal study of over 10,000 current and former cigarette smokers, has been pivotal to these breakthroughs in understanding the genetic basis of COPD. To date, over 20 genetic loci have been convincingly associated with COPD affection status, with additional loci demonstrating association with COPD-related phenotypes such as emphysema, chronic bronchitis, and hypoxemia. In this review, we discuss the contributions of the COPDGene study to the discovery of these genetic associations as well as the ongoing genetic investigations of COPD subtypes, protein biomarkers, and post-genome-wide association study analysis.
Keywords: chronic obstructive pulmonary disease; epidemiology; genetics.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

