Multiple mesenchymal progenitor cell subtypes with distinct functional potential are present within the intimal layer of the hip synovium
- PMID: 30909916
- PMCID: PMC6434889
- DOI: 10.1186/s12891-019-2495-2
Multiple mesenchymal progenitor cell subtypes with distinct functional potential are present within the intimal layer of the hip synovium
Abstract
Background: The synovial membrane adjacent to the articular cartilage is home to synovial mesenchymal progenitor cell (sMPC) populations that have the ability to undergo chondrogenesis. While it has been hypothesized that multiple subtypes of stem and progenitor cells exist in vivo, there is little evidence supporting this hypothesis in human tissues. Furthermore, in most of the published literature on this topic, the cells are cultured before derivation of clonal populations. This gap in the literature makes it difficult to determine if there are distinct MPC subtypes in human synovial tissues, and if so, if these sMPCs express any markers in vivo/in situ that provide information in regards to the function of specific MPC subtypes (e.g. cells with increased chondrogenic capacity)? Therefore, the current study was undertaken to determine if any of the classical MPC cell surface markers provide insight into the differentiation capacity of sMPCs.
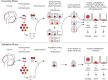
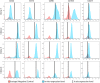
Methods: Clonal populations of sMPCs were derived from a cohort of patients with hip osteoarthritis (OA) and patients at high risk to develop OA using indexed cell sorting. Tri-differentiation potential and cell surface receptor expression of the resultant clones was determined.
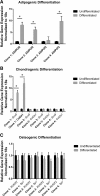
Results: A number of clones with distinct differentiation potential were derived from this cohort, yet the most common cell surface marker profile on MPCs (in situ) that demonstrated chondrogenic potential was determined to be CD90+/CD44+/CD73+. A validation cohort was employed to isolate cells with only this cell surface profile. Isolating cells directly from human synovial tissue with these three markers alone, did not enrich for cells with chondrogenic capacity.
Conclusions: Therefore, additional markers are required to further discriminate the heterogeneous subtypes of MPCs and identify sMPCs with functional properties that are believed to be advantageous for clinical application.
Keywords: Clonal analysis; Heterogeneity; Hip; Osteoarthritis; Synovial progenitor cells.
Conflict of interest statement
Ethics approval and consent to participate
This study protocol was approved by the University of Calgary Human Research Ethics Board (REB15–0005 and REB15–0880). All participants provided written consent to participate. All testing was carried out in accordance with the declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

Similar articles
-
Cell-surface markers identify tissue resident multipotential stem/stromal cell subsets in synovial intimal and sub-intimal compartments with distinct chondrogenic properties.Osteoarthritis Cartilage. 2019 Dec;27(12):1831-1840. doi: 10.1016/j.joca.2019.08.006. Epub 2019 Sep 16. Osteoarthritis Cartilage. 2019. PMID: 31536814
-
Synovial membrane-derived mesenchymal progenitor cells from osteoarthritic joints in dogs possess lower chondrogenic-, and higher osteogenic capacity compared to normal joints.Stem Cell Res Ther. 2022 Sep 5;13(1):457. doi: 10.1186/s13287-022-03144-z. Stem Cell Res Ther. 2022. PMID: 36064441 Free PMC article.
-
Monocyte chemotactic protein-1 inhibits chondrogenesis of synovial mesenchymal progenitor cells: an in vitro study.Stem Cells. 2013 Oct;31(10):2253-65. doi: 10.1002/stem.1477. Stem Cells. 2013. PMID: 23836536
-
Mesenchymal Stem/Progenitor Cells Derived from Articular Cartilage, Synovial Membrane and Synovial Fluid for Cartilage Regeneration: Current Status and Future Perspectives.Stem Cell Rev Rep. 2017 Oct;13(5):575-586. doi: 10.1007/s12015-017-9753-1. Stem Cell Rev Rep. 2017. PMID: 28721683 Review.
-
Synovial membrane mesenchymal stem cells: past life, current situation, and application in bone and joint diseases.Stem Cell Res Ther. 2020 Sep 7;11(1):381. doi: 10.1186/s13287-020-01885-3. Stem Cell Res Ther. 2020. PMID: 32894205 Free PMC article. Review.
Cited by
-
Mesenchymal progenitor cells from non-inflamed versus inflamed synovium post-ACL injury present with distinct phenotypes and cartilage regeneration capacity.Stem Cell Res Ther. 2023 Jun 25;14(1):168. doi: 10.1186/s13287-023-03396-3. Stem Cell Res Ther. 2023. PMID: 37357305 Free PMC article.
-
Synovial fluid mesenchymal progenitor cells from patients with juvenile idiopathic arthritis demonstrate limited self-renewal and chondrogenesis.Sci Rep. 2022 Oct 3;12(1):16530. doi: 10.1038/s41598-022-20880-7. Sci Rep. 2022. PMID: 36192450 Free PMC article.
-
The combinatory effect of sinusoidal electromagnetic field and VEGF promotes osteogenesis and angiogenesis of mesenchymal stem cell-laden PCL/HA implants in a rat subcritical cranial defect.Stem Cell Res Ther. 2019 Dec 16;10(1):379. doi: 10.1186/s13287-019-1464-x. Stem Cell Res Ther. 2019. PMID: 31842985 Free PMC article.
-
What Molecular Recognition Systems Do Mesenchymal Stem Cells/Medicinal Signaling Cells (MSC) Use to Facilitate Cell-Cell and Cell Matrix Interactions? A Review of Evidence and Options.Int J Mol Sci. 2021 Aug 11;22(16):8637. doi: 10.3390/ijms22168637. Int J Mol Sci. 2021. PMID: 34445341 Free PMC article. Review.
-
Addition of High Molecular Weight Hyaluronic Acid to Fibroblast-Like Stromal Cells Modulates Endogenous Hyaluronic Acid Metabolism and Enhances Proteolytic Processing and Secretion of Versican.Cells. 2020 Jul 13;9(7):1681. doi: 10.3390/cells9071681. Cells. 2020. PMID: 32668663 Free PMC article.
References
-
- McIntyre JA, Jones IA, Han B, Vangsness CT. Intra-articular mesenchymal stem cell therapy for the human joint: a systematic review. Am J Sports Med. 2017:036354651773584. 10.1177/0363546517735844. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

