Werner syndrome helicase is a selective vulnerability of microsatellite instability-high tumor cells
- PMID: 30910006
- PMCID: PMC6435321
- DOI: 10.7554/eLife.43333
Werner syndrome helicase is a selective vulnerability of microsatellite instability-high tumor cells
Abstract
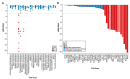
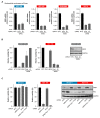
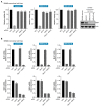
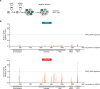
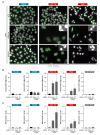
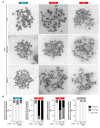
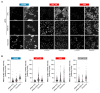
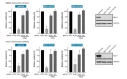
Targeted cancer therapy is based on exploiting selective dependencies of tumor cells. By leveraging recent functional screening data of cancer cell lines we identify Werner syndrome helicase (WRN) as a novel specific vulnerability of microsatellite instability-high (MSI-H) cancer cells. MSI, caused by defective mismatch repair (MMR), occurs frequently in colorectal, endometrial and gastric cancers. We demonstrate that WRN inactivation selectively impairs the viability of MSI-H but not microsatellite stable (MSS) colorectal and endometrial cancer cell lines. In MSI-H cells, WRN loss results in severe genome integrity defects. ATP-binding deficient variants of WRN fail to rescue the viability phenotype of WRN-depleted MSI-H cancer cells. Reconstitution and depletion studies indicate that WRN dependence is not attributable to acute loss of MMR gene function but might arise during sustained MMR-deficiency. Our study suggests that pharmacological inhibition of WRN helicase function represents an opportunity to develop a novel targeted therapy for MSI-H cancers.
Keywords: WRN; cancer biology; colorectal cancer; helicase; human; microsatellite instability; mismatch repair; targeted therapy.
© 2019, Lieb et al.
Conflict of interest statement
SL, SB, EK, JR, CS, KE, AS, AW, JL, GB, RN, NK, MP, MP, SW Full-time employee of Boehringer Ingelheim RCV GmbH & Co KG, Vienna, Austria, KN, Pv, MK, AG, JP No competing interests declared
Figures







Comment in
-
Lethal clues to cancer-cell vulnerability.Nature. 2019 Apr;568(7753):463-464. doi: 10.1038/d41586-019-01086-w. Nature. 2019. PMID: 31000834 No abstract available.
-
Prioritizing synthetic lethal targets with functional genomics.Nat Rev Cancer. 2019 Jun;19(6):305. doi: 10.1038/s41568-019-0147-3. Nat Rev Cancer. 2019. PMID: 31036884 No abstract available.
References
-
- Aggarwal M, Banerjee T, Sommers JA, Iannascoli C, Pichierri P, Shoemaker RH, Brosh RM. Werner syndrome helicase has a critical role in DNA damage responses in the absence of a functional fanconi anemia pathway. Cancer Research. 2013;73:5497–5507. doi: 10.1158/0008-5472.CAN-12-2975. - DOI - PMC - PubMed
-
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jané-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

