Reversing resistance to counter antimicrobial resistance in the World Health Organisation's critical priority of most dangerous pathogens
- PMID: 30910848
- PMCID: PMC6465202
- DOI: 10.1042/BSR20180474
Reversing resistance to counter antimicrobial resistance in the World Health Organisation's critical priority of most dangerous pathogens
Abstract
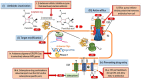
The speed at which bacteria develop antimicrobial resistance far outpace drug discovery and development efforts resulting in untreatable infections. The World Health Organisation recently released a list of pathogens in urgent need for the development of new antimicrobials. The organisms that are listed as the most critical priority are all Gram-negative bacteria resistant to the carbapenem class of antibiotics. Carbapenem resistance in these organisms is typified by intrinsic resistance due to the expression of antibiotic efflux pumps and the permeability barrier presented by the outer membrane, as well as by acquired resistance due to the acquisition of enzymes able to degrade β-lactam antibiotics. In this perspective article we argue the case for reversing resistance by targeting these resistance mechanisms - to increase our arsenal of available antibiotics and drastically reduce antibiotic discovery times - as the most effective way to combat antimicrobial resistance in these high priority pathogens.
Keywords: antibiotic resistance; antibiotics; efflux pump inhibitor; membrane permeability; reversal of resistance; synergism.
© 2019 The Author(s).
Conflict of interest statement
The author declares that there are no competing interests associated with the manuscript.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

