Mutations in NUP160 Are Implicated in Steroid-Resistant Nephrotic Syndrome
- PMID: 30910934
- PMCID: PMC6493979
- DOI: 10.1681/ASN.2018080786
Mutations in NUP160 Are Implicated in Steroid-Resistant Nephrotic Syndrome
Abstract
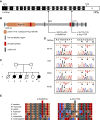
Background: Studies have identified mutations in >50 genes that can lead to monogenic steroid-resistant nephrotic syndrome (SRNS). The NUP160 gene, which encodes one of the protein components of the nuclear pore complex nucleoporin 160 kD (Nup160), is expressed in both human and mouse kidney cells. Knockdown of NUP160 impairs mouse podocytes in cell culture. Recently, siblings with SRNS and proteinuria in a nonconsanguineous family were found to carry compound-heterozygous mutations in NUP160.
Methods: We identified NUP160 mutations by whole-exome and Sanger sequencing of genomic DNA from a young girl with familial SRNS and FSGS who did not carry mutations in other genes known to be associated with SRNS. We performed in vivo functional validation studies on the NUP160 mutations using a Drosophila model.
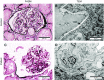
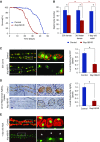
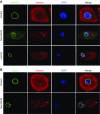
Results: We identified two compound-heterozygous NUP160 mutations, NUP160R1173× and NUP160E803K . We showed that silencing of Drosophila NUP160 specifically in nephrocytes (fly renal cells) led to functional abnormalities, reduced cell size and nuclear volume, and disorganized nuclear membrane structure. These defects were completely rescued by expression of the wild-type human NUP160 gene in nephrocytes. By contrast, expression of the NUP160 mutant allele NUP160R1173× completely failed to rescue nephrocyte phenotypes, and mutant allele NUP160E803K rescued only nuclear pore complex and nuclear lamin localization defects.
Conclusions: Mutations in NUP160 are implicated in SRNS. Our findings indicate that NUP160 should be included in the SRNS diagnostic gene panel to identify additional patients with SRNS and homozygous or compound-heterozygous NUP160 mutations and further strengthen the evidence that NUP160 mutations can cause SRNS.
Keywords: Drosophila; NUP160; genetic renal disease; human genetics; nephrocyte; nephrotic syndrome.
Copyright © 2019 by the American Society of Nephrology.
Figures

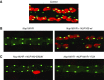
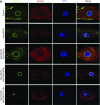

Comment in
-
Filling the Gap: Drosophila Nephrocytes as Model System in Kidney Research.J Am Soc Nephrol. 2019 May;30(5):719-720. doi: 10.1681/ASN.2019020181. Epub 2019 Mar 25. J Am Soc Nephrol. 2019. PMID: 30910933 Free PMC article. No abstract available.
References
-
- Wiggins RC: The spectrum of podocytopathies: A unifying view of glomerular diseases. Kidney Int 71: 1205–1214, 2007 - PubMed
-
- Kestilä M, Lenkkeri U, Männikkö M, Lamerdin J, McCready P, Putaala H, et al. .: Positionally cloned gene for a novel glomerular protein--nephrin--is mutated in congenital nephrotic syndrome. Mol Cell 1: 575–582, 1998 - PubMed
-
- Boute N, Gribouval O, Roselli S, Benessy F, Lee H, Fuchshuber A, et al. .: NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat Genet 24: 349–354, 2000 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

