Anti-Biofilm Activity of Graphene Quantum Dots via Self-Assembly with Bacterial Amyloid Proteins
- PMID: 30912922
- PMCID: PMC6528478
- DOI: 10.1021/acsnano.8b09403
Anti-Biofilm Activity of Graphene Quantum Dots via Self-Assembly with Bacterial Amyloid Proteins
Abstract
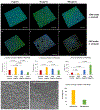
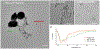
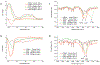
Bacterial biofilms represent an essential part of Earth's ecosystem that can cause multiple ecological, technological, and health problems. The environmental resilience and sophisticated organization of biofilms are enabled by the extracellular matrix that creates a protective network of biomolecules around the bacterial community. Current anti-biofilm agents can interfere with extracellular matrix production but, being based on small molecules, are degraded by bacteria and rapidly diffuse away from biofilms. Both factors severely reduce their efficacy, while their toxicity to higher organisms creates additional barriers to their practicality. In this paper, we report on the ability of graphene quantum dots to effectively disperse mature amyloid-rich Staphylococcus aureus biofilms, interfering with the self-assembly of amyloid fibers, a key structural component of the extracellular matrix. Mimicking peptide-binding biomolecules, graphene quantum dots form supramolecular complexes with phenol-soluble modulins, the peptide monomers of amyloid fibers. Experimental and computational results show that graphene quantum dots efficiently dock near the N-terminus of the peptide and change the secondary structure of phenol-soluble modulins, which disrupts their fibrillation and represents a strategy for mitigation of bacterial communities.
Keywords: biofilm regulation; carbon nanomaterials; extracellular matrix; functional amyloid; nanoscale biomimetics; phenol-soluble modulins; quorum sensing.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures





References
-
- Hill EC; Hill GC Microbial Contamination and Associated Corrosion in Fuels, during Storage, Distribution and Use. Adv. Mater. Res. 2008, 38, 257–268.
-
- Porcelli N; Judd S Chemical Cleaning of Potable Water Membranes: A Review. Sep. Purif. Technol. 2010, 77, 137–143. - PubMed
-
- Costerton JW Bacterial Bofilms: A Common Cause of Persistent Infections. Science (80-.). 1999, 284, 1318–1322. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

