Defined conditions for propagation and manipulation of mouse embryonic stem cells
- PMID: 30914406
- PMCID: PMC6451320
- DOI: 10.1242/dev.173146
Defined conditions for propagation and manipulation of mouse embryonic stem cells
Erratum in
-
Correction: Defined conditions for propagation and manipulation of mouse embryonic stem cells (doi:10.1242/dev.173146).Development. 2019 Apr 11;146(7):dev178970. doi: 10.1242/dev.178970. Development. 2019. PMID: 30975648 Free PMC article. No abstract available.
Abstract
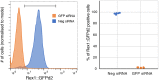
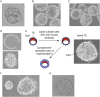
The power of mouse embryonic stem (ES) cells to colonise the developing embryo has revolutionised mammalian developmental genetics and stem cell research. This power is vulnerable, however, to the cell culture environment, deficiencies in which can lead to cellular heterogeneity, adaptive phenotypes, epigenetic aberrations and genetic abnormalities. Here, we provide detailed methodologies for derivation, propagation, genetic modification and primary differentiation of ES cells in 2i or 2i+LIF media without serum or undefined serum substitutes. Implemented diligently, these procedures minimise variability and deviation, thereby improving the efficiency, reproducibility and biological validity of ES cell experimentation.
Keywords: Differentiation; Embryonic stem cells; Pluripotency; Self-renewal.
© 2019. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures
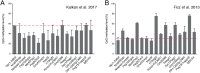

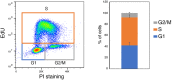

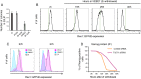
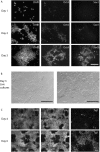
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

