Antagonism of dsRNA-Induced Innate Immune Pathways by NS4a and NS4b Accessory Proteins during MERS Coronavirus Infection
- PMID: 30914508
- PMCID: PMC6437052
- DOI: 10.1128/mBio.00319-19
Antagonism of dsRNA-Induced Innate Immune Pathways by NS4a and NS4b Accessory Proteins during MERS Coronavirus Infection
Abstract
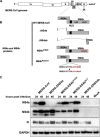
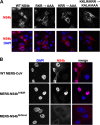
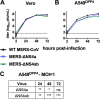
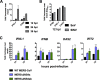
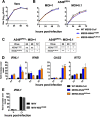
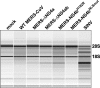
Middle East respiratory syndrome coronavirus (MERS-CoV) was first identified in 2012 as a novel etiological agent of severe respiratory disease in humans. As during infection by other viruses, host sensing of viral double-stranded RNA (dsRNA) induces several antiviral pathways. These include interferon (IFN), oligoadenylate synthetase (OAS)-RNase L, and protein kinase R (PKR). Coronaviruses, including MERS-CoV, potently suppress the activation of these pathways, inducing only modest host responses. Our study describes the functions of two accessory proteins unique to MERS-CoV and related viruses, NS4a and NS4b, during infection in human airway epithelium-derived A549 cells. NS4a has been previously characterized as a dsRNA binding protein, while NS4b is a 2',5'-phosphodiesterase with structural and enzymatic similarity to NS2 encoded by mouse hepatitis virus (MHV). We found that deletion of NS4a results in increased interferon lambda (IFNL1) expression, as does mutation of either the catalytic site or nuclear localization sequence of NS4b. All of the mutant viruses we tested exhibited slight decreases in replication. We previously reported that, like MHV NS2, NS4b antagonizes OAS-RNase L, but suppression of IFN is a previously unidentified function for viral phosphodiesterases. Unexpectedly, deletion of NS4a does not result in robust activation of the PKR or OAS-RNase L pathways. Therefore, MERS-CoV likely encodes other proteins that contribute to suppression or evasion of these antiviral innate immune pathways that should be an important focus of future work. This study provides additional insight into the complex interactions between MERS-CoV and the host immune response.IMPORTANCE Middle East respiratory syndrome coronavirus (MERS-CoV) is the second novel zoonotic coronavirus to emerge in the 21st century and cause outbreaks of severe respiratory disease. More than 2,200 cases and 800 deaths have been reported to date, yet there are no licensed vaccines or treatments. Coronaviruses encode unique accessory proteins that are not required for replication but most likely play roles in immune antagonism and/or pathogenesis. Our study describes the functions of MERS-CoV accessory proteins NS4a and NS4b during infection of a human airway-derived cell line. Loss of these accessory proteins during MERS-CoV infection leads to host antiviral activation and modestly attenuates replication. In the case of both NS4a and NS4b, we have identified roles during infection not previously described, yet the lack of robust activation suggests much remains to be learned about the interactions between MERS-CoV and the infected host.
Keywords: MERS-CoV; coronavirus; interferon antagonist; viral accessory proteins.
Copyright © 2019 Comar et al.
Figures



References
-
- van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus ADME, Haagmans BL, Gorbalenya AE, Snijder EJ, Fouchier RAM. 2012. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 3:e00473-12. doi: 10.1128/mBio.00473-12. - DOI - PMC - PubMed
-
- Reusken CB, Messadi L, Feyisa A, Ularamu H, Godeke G-J, Danmarwa A, Dawo F, Jemli M, Melaku S, Shamaki D, Woma Y, Wungak Y, Gebremedhin EZ, Zutt I, Bosch B-J, Haagmans BL, Koopmans MP. 2014. Geographic distribution of MERS coronavirus among dromedary camels, Africa. Emerg Infect Dis 20:1370–1374. doi: 10.3201/eid2008.140590. - DOI - PMC - PubMed
-
- Wernery U, Rasoul IE, Wong EY, Joseph M, Chen Y, Jose S, Tsang AK, Patteril NAG, Chen H, Elizabeth SK, Yuen K-Y, Joseph S, Xia N, Wernery R, Lau SK, Woo PC. 2015. A phylogenetically distinct Middle East respiratory syndrome coronavirus detected in a dromedary calf from a closed dairy herd in Dubai with rising seroprevalence with age. Emerg Microbes Infect 4:1. doi: 10.1038/emi.2015.74. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
