Comparative Description of the Expression Profile of Interferon-Stimulated Genes in Multiple Cell Lineages Targeted by HIV-1 Infection
- PMID: 30915053
- PMCID: PMC6423081
- DOI: 10.3389/fmicb.2019.00429
Comparative Description of the Expression Profile of Interferon-Stimulated Genes in Multiple Cell Lineages Targeted by HIV-1 Infection
Abstract
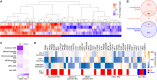
Immediately after viral infections, innate immune sensors recognize viruses and lead to the production of type I interferon (IFN-I). IFN-I upregulates various genes, referred to as IFN-stimulated genes (ISGs), and some ISGs inhibit viral replication. HIV-1, the causative agent of AIDS, mainly infects CD4+ T cells and macrophages and triggers the IFN-I-mediated signaling cascade. Certain ISGs are subsequently upregulated by IFN-I stimulus and potently suppress HIV-1 replication. HIV-1 cell biology has shed light on the molecular understanding of the IFN-I production triggered by HIV-1 infection and the antiviral roles of ISGs. However, the differences in the gene expression patterns following IFN-I stimulus among HIV-1 target cell types are poorly understood. In this study, we hypothesize that the expression profiles of ISGs are different among HIV-1 target cells and address this question by utilizing public transcriptome datasets and bioinformatic techniques. We focus on three cell types intrinsically targeted by HIV-1, including CD4+ T cells, monocytes, and macrophages, and comprehensively compare the expression patterns of ISGs among these cell types. Furthermore, we use the datasets of the differentially expressed genes by HIV-1 infection and the evolutionarily conserved ISGs in mammals and perform comparative transcriptome analyses. We defined 104 'common ISGs' that were upregulated by IFN-I stimulus in CD4+ T cells, monocytes, and macrophages. The ISG expression patterns were different among these three cell types, and intriguingly, both the numbers and the magnitudes of upregulated ISGs by IFN-I stimulus were greatest in macrophages. We also found that the upregulated genes by HIV-1 infection included most 'common ISGs.' Moreover, we determined that the 'common ISGs,' particularly those with antiviral activity, were evolutionarily conserved in mammals. To our knowledge, this study is the first investigation to comprehensively describe (i) the different expression patterns of ISGs among HIV-1 target cells, (ii) the overlap in the genes modulated by IFN-I stimulus and HIV-1 infection and (iii) the evolutionary conservation in mammals of the antiviral ISGs that are expressed in HIV-1 target cells. Our results will be useful for deeply understanding the relationship of the effect of IFN-I and the modulated gene expression by HIV-1 infection.
Keywords: HIV-1; bioinformatics; evolution; interferon-stimulated gene; transcriptomics; type I interferon.
Figures



References
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57 289–300. 10.1111/j.2517-6161.1995.tb02031.x - DOI
LinkOut - more resources
Full Text Sources
Research Materials

