Comparison of Subgingival and Buccal Mucosa Microbiome in Chronic and Aggressive Periodontitis: A Pilot Study
- PMID: 30915280
- PMCID: PMC6421285
- DOI: 10.3389/fcimb.2019.00053
Comparison of Subgingival and Buccal Mucosa Microbiome in Chronic and Aggressive Periodontitis: A Pilot Study
Abstract
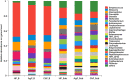
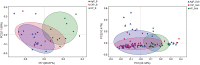
Periodontal microorganisms not only colonize subgingival pockets, but also are detected on various mucous membranes in patients with periodontitis. The object of this pilot study was, using the next-generation sequencing of 16S RNA gene, to characterize the microbiota in two oral habitats (buccal mucosas and subgingival pockets) in patients with different forms of periodontitis. Thirty-two buccal swab samples and 113 subgingival samples were obtained from eleven subjects with chronic periodontitis (ChP), twelve subjects with aggressive periodontitis (AgP), and nine periodontally healthy individuals (HP). Using Miseq Sequencing of 16S rRNA gene, we found that the subgingival and buccal mucosa microbiome of ChP and AgP patients both differed from HP. Meanwhile, Veillonella, Treponema, Filifactor, Fretibacterium, Peptostreptococcaceae_[XI][G-6], Peptostreptococcaceae_[XI][G-5], Bacteroidetes_[G-5], Bacteroidetes_[G-3], Peptostreptococcaceae_[XI][G-4], Peptostreptococcaceae_[XI][G-2] significantly increased both in buccal and subgingival plaque samples in periodontitis subjects (ChP and AgP) compared with HP. Moreover, the results based on the Unweighted UniFrac distance showed that buccal and subgingival plaque samples from the same individuals show higher community divergence than same habitats from different subject samples. This study demonstrated that the microbiome of buccal mucosa can be influenced by periodontitis. However, subgingival and buccal mucosa microbiome seem to be characterized by species-specific colonization patterns. This pilot study provides a glimpse at the changes of subgingival and buccal mucosa associated with periodontitis from a holistic view. Further studies should be taken to illuminate the interplay between these detected changes and periodontitis development.
Keywords: 16S sequencing; aggressive periodontitis; buccal mucosa microbiome; chronic periodontitis; subgingival microbiome.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

