New insights into the structures and interactions of bacterial Y-family DNA polymerases
- PMID: 30916324
- PMCID: PMC6511836
- DOI: 10.1093/nar/gkz198
New insights into the structures and interactions of bacterial Y-family DNA polymerases
Abstract
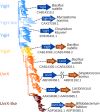
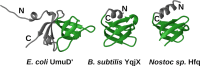
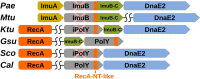
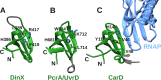
Bacterial Y-family DNA polymerases are usually classified into DinB (Pol IV), UmuC (the catalytic subunit of Pol V) and ImuB, a catalytically dead essential component of the ImuA-ImuB-DnaE2 mutasome. However, the true diversity of Y-family polymerases is unknown. Furthermore, for most of them the structures are unavailable and interactions are poorly characterized. To gain a better understanding of bacterial Y-family DNA polymerases, we performed a detailed computational study. It revealed substantial diversity, far exceeding traditional classification. We found that a large number of subfamilies feature a C-terminal extension next to the common Y-family region. Unexpectedly, in most C-terminal extensions we identified a region homologous to the N-terminal oligomerization motif of RecA. This finding implies a universal mode of interaction between Y-family members and RecA (or ImuA), in the case of Pol V strongly supported by experimental data. In gram-positive bacteria, we identified a putative Pol V counterpart composed of a Y-family polymerase, a YolD homolog and RecA. We also found ImuA-ImuB-DnaE2 variants lacking ImuA, but retaining active or inactive Y-family polymerase, a standalone ImuB C-terminal domain and/or DnaE2. In summary, our analyses revealed that, despite considerable diversity, bacterial Y-family polymerases share previously unanticipated similarities in their structural domains/motifs and interactions.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





Similar articles
-
The RecA-NT homology motif in ImuB mediates the interaction with ImuA', which is essential for DNA damage-induced mutagenesis.J Biol Chem. 2025 Feb;301(2):108108. doi: 10.1016/j.jbc.2024.108108. Epub 2024 Dec 18. J Biol Chem. 2025. PMID: 39706264 Free PMC article.
-
RecA-NT homology motif in ImuB is essential for mycobacterial ImuA'-ImuB protein interaction and mutasome function.bioRxiv [Preprint]. 2023 Mar 28:2023.03.28.534377. doi: 10.1101/2023.03.28.534377. bioRxiv. 2023. Update in: J Biol Chem. 2025 Feb;301(2):108108. doi: 10.1016/j.jbc.2024.108108. PMID: 37034714 Free PMC article. Updated. Preprint.
-
Essential roles for imuA'- and imuB-encoded accessory factors in DnaE2-dependent mutagenesis in Mycobacterium tuberculosis.Proc Natl Acad Sci U S A. 2010 Jul 20;107(29):13093-8. doi: 10.1073/pnas.1002614107. Epub 2010 Jul 6. Proc Natl Acad Sci U S A. 2010. PMID: 20615954 Free PMC article.
-
Escherichia coli Y family DNA polymerases.Front Biosci (Landmark Ed). 2011 Jun 1;16(8):3164-82. doi: 10.2741/3904. Front Biosci (Landmark Ed). 2011. PMID: 21622227 Review.
-
Mutations for Worse or Better: Low-Fidelity DNA Synthesis by SOS DNA Polymerase V Is a Tightly Regulated Double-Edged Sword.Biochemistry. 2016 Apr 26;55(16):2309-18. doi: 10.1021/acs.biochem.6b00117. Epub 2016 Apr 12. Biochemistry. 2016. PMID: 27043933 Free PMC article. Review.
Cited by
-
The translesion polymerase Pol Y1 is a constitutive component of the B. subtilis replication machinery.Nucleic Acids Res. 2024 Sep 9;52(16):9613-9629. doi: 10.1093/nar/gkae637. Nucleic Acids Res. 2024. PMID: 39051562 Free PMC article.
-
Sending out an SOS - the bacterial DNA damage response.Genet Mol Biol. 2022 Oct 10;45(3 Suppl 1):e20220107. doi: 10.1590/1678-4685-GMB-2022-0107. eCollection 2022. Genet Mol Biol. 2022. PMID: 36288458 Free PMC article.
-
A Comprehensive View of Translesion Synthesis in Escherichia coli.Microbiol Mol Biol Rev. 2020 Jun 17;84(3):e00002-20. doi: 10.1128/MMBR.00002-20. Print 2020 Aug 19. Microbiol Mol Biol Rev. 2020. PMID: 32554755 Free PMC article. Review.
-
pLS20 is the archetype of a new family of conjugative plasmids harboured by Bacillus species.NAR Genom Bioinform. 2021 Oct 27;3(4):lqab096. doi: 10.1093/nargab/lqab096. eCollection 2021 Dec. NAR Genom Bioinform. 2021. PMID: 34729475 Free PMC article.
-
DNA polymerase swapping in Caudoviricetes bacteriophages.Virol J. 2024 Aug 26;21(1):200. doi: 10.1186/s12985-024-02482-z. Virol J. 2024. PMID: 39187833 Free PMC article.
References
-
- Kornberg A., Baker T.A.. Kornberg A, Baker T. DNA replication. 1992; 2nd ed. NY: W.H. Freeman.
-
- Ohmori H., Friedberg E.C., Fuchs R.P., Goodman M.F., Hanaoka F., Hinkle D., Kunkel T.A., Lawrence C.W., Livneh Z., Nohmi T. et al. .. The Y-family of DNA polymerases. Mol. Cell. 2001; 8:7–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

