Transcriptional and post-transcriptional events trigger de novo infB expression in cold stressed Escherichia coli
- PMID: 30916329
- PMCID: PMC6511841
- DOI: 10.1093/nar/gkz187
Transcriptional and post-transcriptional events trigger de novo infB expression in cold stressed Escherichia coli
Abstract
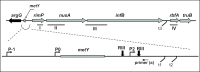
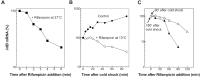
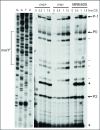
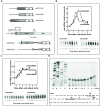
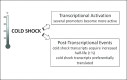
After a 37 to 10°C temperature downshift the level of translation initiation factor IF2, like that of IF1 and IF3, increases at least 3-fold with respect to the ribosomes. To clarify the mechanisms and conditions leading to cold-stress induction of infB expression, the consequences of this temperature shift on infB (IF2) transcription, infB mRNA stability and translation were analysed. The Escherichia coli gene encoding IF2 is part of the metY-nusA-infB operon that contains three known promoters (P-1, P0 and P2) in addition to two promoters P3 and P4 identified in this study, the latter committed to the synthesis of a monocistronic mRNA encoding exclusively IF2. The results obtained indicate that the increased level of IF2 following cold stress depends on three mechanisms: (i) activation of all the promoters of the operon, P-1 being the most cold-responsive, as a likely consequence of the reduction of the ppGpp level that follows cold stress; (ii) a large increase in infB mRNA half-life and (iii) the cold-shock induced translational bias that ensures efficient translation of infB mRNA by the translational apparatus of cold shocked cells. A comparison of the mechanisms responsible for the cold shock induction of the three initiation factors is also presented.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
Translation initiation factor IF2 contributes to ribosome assembly and maturation during cold adaptation.Nucleic Acids Res. 2019 May 21;47(9):4652-4662. doi: 10.1093/nar/gkz188. Nucleic Acids Res. 2019. PMID: 30916323 Free PMC article.
-
Cold-shock-induced de novo transcription and translation of infA and role of IF1 during cold adaptation.Mol Microbiol. 2007 May;64(3):807-21. doi: 10.1111/j.1365-2958.2007.05699.x. Mol Microbiol. 2007. PMID: 17462025
-
Structural requirements of the mRNA for intracistronic translation initiation of the enterobacterial infB gene.Genes Cells. 2002 Sep;7(9):901-10. doi: 10.1046/j.1365-2443.2002.00571.x. Genes Cells. 2002. PMID: 12296821
-
Transcriptional and post-transcriptional control of cold-shock genes.J Mol Biol. 2003 Aug 15;331(3):527-39. doi: 10.1016/s0022-2836(03)00732-0. J Mol Biol. 2003. PMID: 12899826 Review.
-
Cold Shock Response in Bacteria.Annu Rev Genet. 2021 Nov 23;55:377-400. doi: 10.1146/annurev-genet-071819-031654. Epub 2021 Sep 16. Annu Rev Genet. 2021. PMID: 34530639 Review.
Cited by
-
RNase-mediated reprogramming of Yersinia virulence.PLoS Pathog. 2024 Aug 19;20(8):e1011965. doi: 10.1371/journal.ppat.1011965. eCollection 2024 Aug. PLoS Pathog. 2024. PMID: 39159284 Free PMC article.
-
Emerging Adaptive Strategies Under Temperature Fluctuations in a Laboratory Evolution Experiment of Escherichia Coli.Front Microbiol. 2021 Oct 22;12:724982. doi: 10.3389/fmicb.2021.724982. eCollection 2021. Front Microbiol. 2021. PMID: 34745030 Free PMC article.
-
RNase III, Ribosome Biogenesis and Beyond.Microorganisms. 2021 Dec 17;9(12):2608. doi: 10.3390/microorganisms9122608. Microorganisms. 2021. PMID: 34946208 Free PMC article. Review.
-
The Ribosome as a Switchboard for Bacterial Stress Response.Front Microbiol. 2021 Jan 8;11:619038. doi: 10.3389/fmicb.2020.619038. eCollection 2020. Front Microbiol. 2021. PMID: 33584583 Free PMC article. Review.
-
Cold stress enhances cryotolerance in Lacticaseibacillus rhamnosus B6 via membrane lipid remodeling and differential protein expression.Curr Res Microb Sci. 2025 Aug 5;9:100453. doi: 10.1016/j.crmicr.2025.100453. eCollection 2025. Curr Res Microb Sci. 2025. PMID: 40837524 Free PMC article.
References
-
- Thieringer H.A., Jones P.G., Inouye M.. Cold shock and adaptation. Bioessays. 1998; 20:49–57. - PubMed
-
- Yamanaka K. Cold shock response in Escherichia coli. J. Mol. Microbiol. Biotechnol. 1999; 1:193–202. - PubMed
-
- Graumann P., Wendrich T.M., Weber M.H., Schröder K., Marahiel M.A.. A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol. Microbiol. 1997; 25:741–756. - PubMed
-
- Gualerzi C.O., Giuliodori A.M., Pon C.L.. Transcriptional and post-transcriptional control of cold-shock genes. J. Mol. Biol. 2003; 331:527–539. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

