Antigen receptor control of methionine metabolism in T cells
- PMID: 30916644
- PMCID: PMC6497464
- DOI: 10.7554/eLife.44210
Antigen receptor control of methionine metabolism in T cells
Abstract
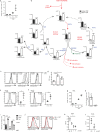
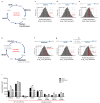
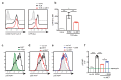
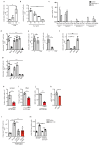
Immune activated T lymphocytes modulate the activity of key metabolic pathways to support the transcriptional reprograming and reshaping of cell proteomes that permits effector T cell differentiation. The present study uses high resolution mass spectrometry and metabolic labelling to explore how murine T cells control the methionine cycle to produce methyl donors for protein and nucleotide methylations. We show that antigen receptor engagement controls flux through the methionine cycle and RNA and histone methylations. We establish that the main rate limiting step for protein synthesis and the methionine cycle is control of methionine transporter expression. Only T cells that respond to antigen to upregulate and sustain methionine transport are supplied with methyl donors that permit the dynamic nucleotide methylations and epigenetic reprogramming that drives T cell differentiation. These data highlight how the regulation of methionine transport licenses use of methionine for multiple fundamental processes that drive T lymphocyte proliferation and differentiation.
Keywords: T cell activation; immunology; inflammation; lymphocyte; methionine metabolism; mouse; nutrient uptake; proteomics.
© 2019, Sinclair et al.
Conflict of interest statement
LS, AH, AB, LS, JH, AM, XL, ST, PT, JR, JL, AL, DC No competing interests declared
Figures



Comment in
- doi: 10.7554/eLife.47221
References
-
- Baird FE, Bett KJ, MacLean C, Tee AR, Hundal HS, Taylor PM. Tertiary active transport of amino acids reconstituted by coexpression of system A and L transporters in Xenopus oocytes. American Journal of Physiology-Endocrinology and Metabolism. 2009;297:E822–E829. doi: 10.1152/ajpendo.00330.2009. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 205023/Z/16/Z (Principal Research Fellowship)/WT_/Wellcome Trust/United Kingdom
- 205023/Z/16/Z/Wellcome/International
- P30 DK058404/DK/NIDDK NIH HHS/United States
- 097418/Z/11/Z/Wellcome/International
- 105024/Z/14/Z (Wellcome Trust Strategic Award)/WT_/Wellcome Trust/United Kingdom
- 202950/Z/16/Z/Wellcome/International
- WT_/Wellcome Trust/United Kingdom
- R01 CA217987/CA/NCI NIH HHS/United States
- R01 DK105550/DK/NIDDK NIH HHS/United States
- 202950/Z/16/Z (Wellcome Trust Equipment Award)/WT_/Wellcome Trust/United Kingdom
- R01 HL136664/HL/NHLBI NIH HHS/United States
- S10 OD018164/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

