The NMR-based characterization of the FTY720-SET complex reveals an alternative mechanism for the attenuation of the inhibitory SET-PP2A interaction
- PMID: 30917007
- PMCID: PMC6529350
- DOI: 10.1096/fj.201802264R
The NMR-based characterization of the FTY720-SET complex reveals an alternative mechanism for the attenuation of the inhibitory SET-PP2A interaction
Abstract
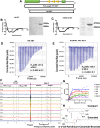
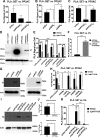
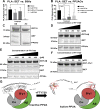
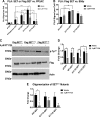
The su(var)3-9, enhancer of zeste, trithorax (SET)/inhibitor 2 of protein phosphatase 2A (PP2A) oncoprotein binds and inhibits PP2A, composed of various isoforms of scaffolding, regulatory, and catalytic subunits. Targeting SET with a sphingolipid analog drug fingolimod (FTY720) or ceramide leads to the reactivation of tumor suppressor PP2A. However, molecular details of the SET-FTY720 or SET-ceramide, and mechanism of FTY720-dependent PP2A activation, remain unknown. Here, we report the first in solution examination of the SET-FTY720 or SET-ceramide complexes by NMR spectroscopy. FTY720-ceramide binding resulted in chemical shifts of residues residing at the N terminus of SET, preventing its dimerization or oligomerization. This then released SET from PP2ACα, resulting in PP2A activation, while monomeric SET remained associated with the B56γ. Our data also suggest that the PP2A holoenzyme, composed of PP2A-Aβ, PP2A-B56γ, and PP2ACα subunits, is selectively activated in response to the formation of the SET-FTY720 complex in A549 cells. Various PP2A-associated downstream effector proteins in the presence or absence of FTY720 were then identified by stable isotope labeling with amino cells in cell culture, including tumor suppressor nonmuscle myosin IIA. Attenuation of FTY720-SET association by point mutations of residues that are involved in FTY720 binding or dephosphorylation of SET at Serine 171, enhanced SET oligomerization and the formation of the SET-PP2A inhibitory complex, leading to resistance to FTY720-dependent PP2A activation.-De Palma, R. M., Parnham, S. R., Li, Y., Oaks, J. J., Peterson, Y. K., Szulc, Z. M., Roth, B. M., Xing, Y., Ogretmen, B. The NMR-based characterization of the FTY720-SET complex reveals an alternative mechanism for the attenuation of the inhibitory SET-PP2A interaction.
Keywords: ceramide; fingolimod; lipid-protein binding; sphingolipid.
Conflict of interest statement
The authors thank Dr. Aiping Bai and Mr. Jason Pierce for help in ceramide measurements at the Lipidomics Shared Resource facility, Department of Biochemistry and Molecular Biology, Medical University of South Carolina. The authors also thank Mr. Brett Bechtol (Hollings Cancer Center, Medical University of South Carolina) for editorial review and assistance. The authors thank the members of the B.O. laboratory for technical support and helpful discussions. The authors also thank Dr. Do-Sung Kim (Department of Biochemistry and Molecular Biology, Hollings Cancer Center, Medical University of South Carolina) for help and assistance in our experimental procedures. This work is supported by research funding from the U.S. National Institutes of Health (NIH) National Cancer Institute (CA088932, CA173687, CA214461, and P01 CA203628, to B.O.), and the NIH National Institute of Dental and Craniofacial Research (DE016572, to B.O.). The core facilities utilized were constructed using support from NIH National Center for Research Resources (C06 RR015455), Hollings Cancer Center Support Grant (P30 CA138313), or Center of Biomedical Research Excellence (CoBRE) in Lipidomics and Pathobiology (P30 GM103339). B.O. is the founder and Chief Scientific Officer of Lipo-Immuno Tech, LLC., and Z.M.S. is the director of synthetic chemistry of the Lipo-Immuno Tech. The authors declare no conflicts of interest.
Figures




References
-
- Virshup D. M. (2000) Protein phosphatase 2A: a panoply of enzymes. Curr. Opin. Cell Biol. 12, 180–185 - PubMed
-
- Herzog F., Kahraman A., Boehringer D., Mak R., Bracher A., Walzthoeni T., Leitner A., Beck M., Hartl F.-U., Ban N., Malmström L., Aebersold R. (2012) Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science 337, 1348–1352 - PubMed
-
- Saydam G., Aydin H. H., Sahin F., Selvi N., Oktem G., Terzioglu E., Buyukkececi F., Omay S. B. (2003) Involvement of protein phosphatase 2A in interferon-alpha-2b-induced apoptosis in K562 human chronic myelogenous leukaemia cells. Leuk. Res. 27, 709–717 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

