Primary endothelial cell-specific regulation of hypoxia-inducible factor (HIF)-1 and HIF-2 and their target gene expression profiles during hypoxia
- PMID: 30917010
- PMCID: PMC6593883
- DOI: 10.1096/fj.201802650RR
Primary endothelial cell-specific regulation of hypoxia-inducible factor (HIF)-1 and HIF-2 and their target gene expression profiles during hypoxia
Abstract
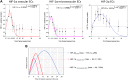
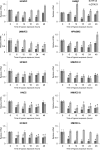
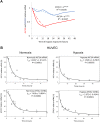
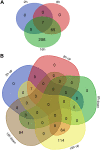
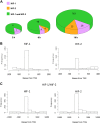
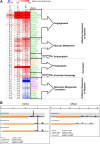
During hypoxia, a cellular adaptive response activates hypoxia-inducible factors (HIFs; HIF-1 and HIF-2) that respond to low tissue-oxygen levels and induce the expression of a number of genes that promote angiogenesis, energy metabolism, and cell survival. HIF-1 and HIF-2 regulate endothelial cell (EC) adaptation by activating gene-signaling cascades that promote endothelial migration, growth, and differentiation. An HIF-1 to HIF-2 transition or switch governs this process from acute to prolonged hypoxia. In the present study, we evaluated the mechanisms governing the HIF switch in 10 different primary human ECs from different vascular beds during the early stages of hypoxia. The studies demonstrate that the switch from HIF-1 to HIF-2 constitutes a universal mechanism of cellular adaptation to hypoxic stress and that HIF1A and HIF2A mRNA stability differences contribute to HIF switch. Furthermore, using 4 genome-wide mRNA expression arrays of HUVECs during normoxia and after 2, 8, and 16 h of hypoxia, we show using bioinformatics analyses that, although a number of genes appeared to be regulated exclusively by HIF-1 or HIF-2, the largest number of genes appeared to be regulated by both.-Bartoszewski, R., Moszyńska, A., Serocki, M., Cabaj, A., Polten, A., Ochocka, R., Dell'Italia, L., Bartoszewska, S., Króliczewski, J., Dąbrowski, M., Collawn, J. F. Primary endothelial cell-specific regulation of hypoxia-inducible factor (HIF)-1 and HIF-2 and their target gene expression profiles during hypoxia.
Keywords: human endothelial cells.
Conflict of interest statement
The authors thank Perlan Technologies, Polska Spółka z ograniczoną odpowiedzialnością (Sp. z o.o.) for performing microarray experiments and analysis. This research was funded by National Science Center SONATA BIS Program under contract UMO-2015/18/E/NZ3/00687 (to R.B.). J.F.C. was funded by the U.S. National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases Grant P30 DK072482, and a Research Development Program grant from the Cystic Fibrosis Foundation. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results. The authors declare no conflicts of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

