Simplified Intestinal Microbiota to Study Microbe-Diet-Host Interactions in a Mouse Model
- PMID: 30917328
- PMCID: PMC6444000
- DOI: 10.1016/j.celrep.2019.02.090
Simplified Intestinal Microbiota to Study Microbe-Diet-Host Interactions in a Mouse Model
Abstract
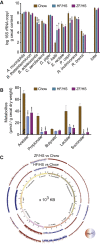
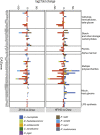
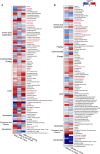
The gut microbiota can modulate human metabolism through interactions with macronutrients. However, microbiota-diet-host interactions are difficult to study because bacteria interact in complex food webs in concert with the host, and many of the bacteria are not yet characterized. To reduce the complexity, we colonize mice with a simplified intestinal microbiota (SIM) composed of ten sequenced strains isolated from the human gut with complementing pathways to metabolize dietary fibers. We feed the SIM mice one of three diets (chow [fiber rich], high-fat/high-sucrose, or zero-fat/high-sucrose diets [both low in fiber]) and investigate (1) how dietary fiber, saturated fat, and sucrose affect the abundance and transcriptome of the SIM community, (2) the effect of microbe-diet interactions on circulating metabolites, and (3) how microbiota-diet interactions affect host metabolism. Our SIM model can be used in future studies to help clarify how microbiota-diet interactions contribute to metabolic diseases.
Keywords: diet; metabolome; microbiota; transcriptome.
Crown Copyright © 2019. Published by Elsevier Inc. All rights reserved.
Figures



Comment in
-
Diet influences microbe-host interaction.Nat Methods. 2019 May;16(5):361. doi: 10.1038/s41592-019-0413-z. Nat Methods. 2019. PMID: 31040430 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

