Analysis of 50 Neurodegenerative Genes in Clinically Diagnosed Early-Onset Alzheimer's Disease
- PMID: 30917570
- PMCID: PMC6471359
- DOI: 10.3390/ijms20061514
Analysis of 50 Neurodegenerative Genes in Clinically Diagnosed Early-Onset Alzheimer's Disease
Abstract
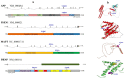
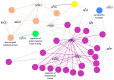
Alzheimer's disease (AD), Parkinson's disease (PD), frontotemporal dementia (FTD), amyotrophic lateral sclerosis (ALS), Huntington's disease (HD), and prion diseases have a certain degree of clinical, pathological, and molecular overlapping. Previous studies revealed that many causative mutations in AD, PD, and FTD/ALS genes could be found in clinical familial and sporadic AD. To further elucidate the missing heritability in early-onset Alzheimer's disease (EOAD), we genetically characterized a Thai EOAD cohort by Next-Generation Sequencing (NGS) with a high depth of coverage, capturing variants in 50 previously recognized AD and other related disorders' genes. A novel mutation, APP p.V604M, and the known causative variant, PSEN1 p.E184G, were found in two of the familiar cases. Remarkably, among 61 missense variants were additionally discovered from 21 genes out of 50 genes, six potential mutations including MAPT P513A, LRRK2 p.R1628P, TREM2 p.L211P, and CSF1R (p.P54Q and pL536V) may be considered to be probably/possibly pathogenic and risk factors for other dementia leading to neuronal degeneration. All allele frequencies of the identified missense mutations were compared to 622 control individuals. Our study provides initial evidence that AD and other neurodegenerative diseases may represent shades of the same disease spectrum, and consideration should be given to offer exactly embracing genetic testing to patients diagnosed with EOAD. Our results need to be further confirmed with a larger cohort from this area.
Keywords: 50 genes; Alzheimer’s disease; EOAD; Thailand; next generation sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Giau V.V., Bagyinszky E., An S.S.A., Kim S. Clinical genetic strategies for early onset neurodegenerative diseases. Mol. Cell. Toxicol. 2018;14:123–142. doi: 10.1007/s13273-018-0015-3. - DOI
-
- Hagenaars S.P., Radaković R., Crockford C., Fawns-Ritchie C., International FTD-Genomics Consortium. Harris S.E., Gale C.R., Deary I.J. Genetic risk for neurodegenerative disorders, and its overlap with cognitive ability and physical function. PLoS ONE. 2018;13:e0198187. doi: 10.1371/journal.pone.0198187. - DOI - PMC - PubMed
-
- Van Giau V., An S.S.A., Bagyinszky E., Kim S. Gene panels and primers for next generation sequencing studies on neurodegenerative disorders. Mol. Cell. Toxicol. 2015;11:89–143. doi: 10.1007/s13273-015-0011-9. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

