Molecular Typing of Mycoplasma pneumoniae Strains in Sweden from 1996 to 2017 and the Emergence of a New P1 Cytadhesin Gene, Variant 2e
- PMID: 30918047
- PMCID: PMC6535615
- DOI: 10.1128/JCM.00049-19
Molecular Typing of Mycoplasma pneumoniae Strains in Sweden from 1996 to 2017 and the Emergence of a New P1 Cytadhesin Gene, Variant 2e
Abstract
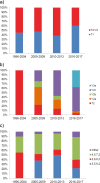
Mycoplasma pneumoniae causes respiratory infections, such as community-acquired pneumonia (CAP), with epidemics recurring every 3 to 7 years. In 2010 and 2011, many countries experienced an extraordinary epidemic peak. The cause of these recurring epidemics is not understood, but decreasing herd immunity and shifts in the strains' antigenic properties have been suggested as contributing factors. M. pneumoniae PCR-positive samples were collected between 1996 and 2017 from four neighboring counties inhabited by 12% of Sweden's population. A total of 578 isolates were characterized directly from 624 clinical samples using P1 typing by sequencing and multilocus variable number tandem repeat analysis (MLVA). A fluorescence resonance energy transfer (FRET)-PCR approach was also used to detect mutations associated with macrolide resistance in the 23S rRNA gene. Through P1 typing, the strains were classified into type 1 and type 2, as well as variants 2a, 2b, 2c, and a new variant found in nine of the strains, denoted variant 2e. Twelve MLVA types were distinguished, and 3-5-6-2 (42.4%), 4-5-7-2 (37.4%), and 3-6-6-2 (14.9%) predominated. Several P1 and MLVA types cocirculated each year, but type 2/variant 2 strains and MLVA types 3-5-6-2 and 4-5-7-2 predominated during the epidemic period comprising the peak of 2010 and 2011. In 2016 and 2017, type 1 became more common, and MLVA type 4-5-7-2 predominated. We also found that 0.2% (1/578) of the strains carried a macrolide resistance-associated mutation, indicating a very low prevalence of macrolide resistance in this region of Sweden.
Keywords: MLVA; Mycoplasma pneumoniae; P1 typing; molecular typing.
Copyright © 2019 Gullsby et al.
Figures



References
-
- Diaz MH, Desai HP, Morrison SS, Benitez AJ, Wolff BJ, Caravas J, Read TD, Dean D, Winchell JM. 2017. Comprehensive bioinformatics analysis of Mycoplasma pneumoniae genomes to investigate underlying population structure and type-specific determinants. PLoS One 12:e0174701. doi:10.1371/journal.pone.0174701. - DOI - PMC - PubMed
-
- Matsuoka M, Narita M, Okazaki N, Ohya H, Yamazaki T, Ouchi K, Suzuki I, Andoh T, Kenri T, Sasaki Y, Horino A, Shintani M, Arakawa Y, Sasaki T. 2004. Characterization and molecular analysis of macrolide-resistant Mycoplasma pneumoniae clinical isolates obtained in Japan. Antimicrob Agents Chemother 48:4624–4630. doi:10.1128/AAC.48.12.4624-4630.2004. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

