Parent of origin genetic effects on methylation in humans are common and influence complex trait variation
- PMID: 30918249
- PMCID: PMC6437195
- DOI: 10.1038/s41467-019-09301-y
Parent of origin genetic effects on methylation in humans are common and influence complex trait variation
Erratum in
-
Publisher Correction: Parent of origin genetic effects on methylation in humans are common and influence complex trait variation.Nat Commun. 2019 May 1;10(1):2069. doi: 10.1038/s41467-019-10155-7. Nat Commun. 2019. PMID: 31043600 Free PMC article.
Abstract
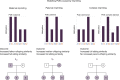
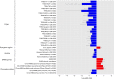
Parent-of-origin effects (POE) exist when there is differential expression of alleles inherited from the two parents. A genome-wide scan for POE on DNA methylation at 639,238 CpGs in 5,101 individuals identifies 733 independent methylation CpGs potentially influenced by POE at a false discovery rate ≤ 0.05 of which 331 had not previously been identified. Cis and trans methylation quantitative trait loci (mQTL) regulate methylation variation through POE at 54% (399/733) of the identified POE-influenced CpGs. The combined results provide strong evidence for previously unidentified POE-influenced CpGs at 171 independent loci. Methylation variation at 14 of the POE-influenced CpGs is associated with multiple metabolic traits. A phenome-wide association analysis using the POE mQTL SNPs identifies a previously unidentified imprinted locus associated with waist circumference. These results provide a high resolution population-level map for POE on DNA methylation sites, their local and distant regulators and potential consequences for complex traits.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

