Influence of p53 Isoform Expression on Survival in High-Grade Serous Ovarian Cancers
- PMID: 30918304
- PMCID: PMC6437169
- DOI: 10.1038/s41598-019-41706-z
Influence of p53 Isoform Expression on Survival in High-Grade Serous Ovarian Cancers
Abstract
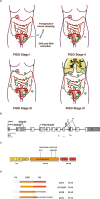
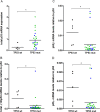
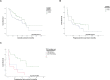
High-grade serous ovarian carcinoma (HGSOC) is characterised by alterations in the p53 pathway. The expression levels of p53 isoforms have been shown to be associated with patient survival in several cancers. This study examined the predictive and prognostic effects of the expression levels of TP53 pre-mRNA splicing isoforms and TP53 mutations in tumour tissues in 40 chemotherapy responders and 29 non-responders with HGSOC. The mRNA expression levels from total p53, and total Δ133p53, p53β, p53γ isoforms were determined by RT-qPCR, and TP53 mutation status by targeted massive parallel sequencing. The results from these analyses were correlated with the clinical outcome parameters. No differential expression of p53 isoforms could be detected between the chemosensitive and chemoresistant subgroups. In a multivariate Cox regression model, high levels of total Δ133p53 were found to be an independent prognosticator for improved overall survival (HR = 0.422, p = 0.018, 95% CI: 0.207-0.861) and reached borderline significance for progression-free survival (HR = 0.569, p = 0.061, 95% CI: 0.315-1.027). TP53 mutations resulting in loss of function or located at known hotspots were predictive of tumour characteristics and disease progression. These findings suggest that total Δ133p53 mRNA can be a biomarker for survival in HGSOC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

