Syndecan 1 is a critical mediator of macropinocytosis in pancreatic cancer
- PMID: 30918400
- PMCID: PMC6661074
- DOI: 10.1038/s41586-019-1062-1
Syndecan 1 is a critical mediator of macropinocytosis in pancreatic cancer
Abstract
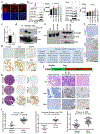
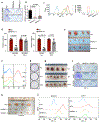
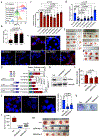
Pancreatic ductal adenocarcinoma (PDAC) remains recalcitrant to all forms of cancer treatment and carries a five-year survival rate of only 8%1. Inhibition of oncogenic KRAS (hereafter KRAS*), the earliest lesion in disease development that is present in more than 90% of PDACs, and its signalling surrogates has yielded encouraging preclinical results with experimental agents2-4. However, KRAS*-independent disease recurrence following genetic extinction of Kras* in mouse models anticipates the need for co-extinction strategies5,6. Multiple oncogenic processes are initiated at the cell surface, where KRAS* physically and functionally interacts to direct signalling that is essential for malignant transformation and tumour maintenance. Insights into the complexity of the functional cell-surface-protein repertoire (surfaceome) have been technologically limited until recently and-in the case of PDAC-the genetic control of the function and composition of the PDAC surfaceome in the context of KRAS* signalling remains largely unknown. Here we develop an unbiased, functional target-discovery platform to query KRAS*-dependent changes of the PDAC surfaceome, which reveals syndecan 1 (SDC1, also known as CD138) as a protein that is upregulated at the cell surface by KRAS*. Localization of SDC1 at the cell surface-where it regulates macropinocytosis, an essential metabolic pathway that fuels PDAC cell growth-is essential for disease maintenance and progression. Thus, our study forges a mechanistic link between KRAS* signalling and a targetable molecule driving nutrient salvage pathways in PDAC and validates oncogene-driven surfaceome annotation as a strategy to identify cancer-specific vulnerabilities.
Figures











References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

