Molecular architecture of the zebra finch arcopallium
- PMID: 30919954
- PMCID: PMC6879308
- DOI: 10.1002/cne.24688
Molecular architecture of the zebra finch arcopallium
Abstract
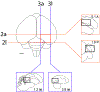
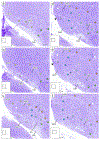
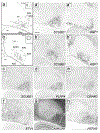
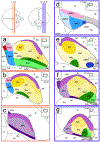
The arcopallium, a key avian forebrain region, receives inputs from numerous brain areas and is a major source of descending sensory and motor projections. While there is evidence of arcopallial subdivisions, the internal organization or the arcopallium is not well understood. The arcopallium is also considered the avian homologue of mammalian deep cortical layers and/or amygdalar subdivisions, but one-to-one correspondences are controversial. Here we present a molecular characterization of the arcopallium in the zebra finch, a passerine songbird species and a major model organism for vocal learning studies. Based on in situ hybridization for arcopallial-expressed transcripts (AQP1, C1QL3, CBLN2, CNTN4, CYP19A1, ESR1/2, FEZF2, MGP, NECAB2, PCP4, PVALB, SCN3B, SCUBE1, ZBTB20, and others) in comparison with cytoarchitectonic features, we have defined 20 distinct regions that can be grouped into six major domains (anterior, posterior, dorsal, ventral, medial, and intermediate arcopallium, respectively; AA, AP, AD, AV, AM, and AI). The data also help to establish the arcopallium as primarily pallial, support a unique topography of the arcopallium in passerines, highlight similarities between the vocal robust nucleus of the arcopallium (RA) and AI, and provide insights into the similarities and differences of cortical and amygdalar regions between birds and mammals. We also propose the use of AMV (instead of nucleus taenia/TnA), AMD, AD, and AI as initial steps toward a universal arcopallial nomenclature. Besides clarifying the internal organization of the arcopallium, the data provide a coherent basis for further functional and comparative studies of this complex avian brain region.
Keywords: RRID: SCR_012988); ZEBrA (www.zebrafinchatlas.org); amygdala; avian; cortical layers; in situ hybridization; molecularly defined brain regions; oscine songbird; taenia.
© 2019 Wiley Periodicals, Inc.
Figures









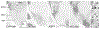








References
-
- Abellan A, Legaz I, Vernier B, Retaux S, Medina L. 2009. Olfactory and amygdalar structures of the chicken ventral pallium based on the combinatorial expression patterns of LIM and other developmental regulatory genes. J Comp Neurol 516(3):166–186. - PubMed
-
- Absil P, Braquenier JB, Balthazart J, Ball GF. 2002. Effects of lesions of nucleus taeniae on appetitive and consummatory aspects of male sexual behavior in Japanese quail. Brain Behav Evol 60(1):13–35. - PubMed
-
- Acharya KD, Veney SL. 2012. Characterization of the G-protein-coupled membrane-bound estrogen receptor GPR30 in the zebra finch brain reveals a sex difference in gene and protein expression. Dev Neurobiol 72(11):1433–1446. - PubMed
-
- Arnold AP. 1980. Quantitative analysis of sex differences in hormone accumulation in the zebra finch brain: methodological and theoretical issues. J Comp Neurol 189(3):421–436. - PubMed
-
- Atoji Y, Saito S, Wild JM. 2006. Fiber connections of the compact division of the posterior pallial amygdala and lateral part of the bed nucleus of the stria terminalis in the pigeon (Columba livia). J Comp Neurol 499(2):161–182. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

