miRNAs expression of oral squamous cell carcinoma patients: Validation of two putative biomarkers
- PMID: 30921188
- PMCID: PMC6456104
- DOI: 10.1097/MD.0000000000014922
miRNAs expression of oral squamous cell carcinoma patients: Validation of two putative biomarkers
Abstract
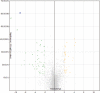
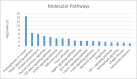
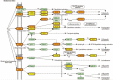
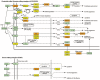
microRNA expression patterns have provided new directions in the search of biomarkers with prognostic value and even in the search of novel therapeutic targets for several neoplasms. Specifically, miRNAs profiling in oral squamous cell carcinoma (OSCC) represents a web of intrigue in the study of oral carcinogenesis. The objective of the present study was twofold:The first study phase comprised case-control groups: A) 8 OSCC-affected patients and 8 healthy controls. Microarray technology (Affymetrix miRNA Array Plate 4.1) was used for miRNAs expression profile. Deregulated miRNAs were studied using Diana Tools miRPath 3.0 to associate miRNA targets with molecular pathways via Kyoto Encyclopedia of Genes and Genomes (KEGG). In a second phase, 2 miRNAs chosen for the subsequent RT-qPCR validation were studied in a second OSSC cohort (n = 8).Microarray analysis identified 80 deregulated miRNAs (35 over-expressed and 45 under-expressed). Two miRNAs (miR-497-5p and miR-4417) were chosen for further validation via RT-qPCR. Prognostic analysis did not ascertain relevant relation between miR-497-5p or miR-4417 expression and clinical or pathological parameters, except high miR-4417 in the case of nodular affectation (P = .035) and diminished miR-497-5p radiotherapy-treated patients (P = .05). KEGG analysis revealed that deregulated miRNAs were implicated in several biological pathways such as Proteoglycans in cancer.Our data suggest an altered miRNAs profiling in OSCC-affected patients. We have verified the altered expression of miR-497-5p and miR-4417 in OSCC samples and related the deregulated miRNAs with the 'proteoglycans in cancer' pathway. Further longitudinal studies with large samples are warranted to confirm the present findings.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures
Similar articles
-
Oral squamous cell carcinoma: microRNA expression profiling and integrative analyses for elucidation of tumourigenesis mechanism.Mol Cancer. 2016 Apr 7;15:28. doi: 10.1186/s12943-016-0512-8. Mol Cancer. 2016. PMID: 27056547 Free PMC article.
-
Genome-wide study of salivary miRNAs identifies miR-423-5p as promising diagnostic and prognostic biomarker in oral squamous cell carcinoma.Theranostics. 2021 Jan 1;11(6):2987-2999. doi: 10.7150/thno.45157. eCollection 2021. Theranostics. 2021. PMID: 33456584 Free PMC article.
-
Circulating miRNAs as biomarkers for oral squamous cell carcinoma recurrence in operated patients.Oncotarget. 2017 Jan 31;8(5):8206-8214. doi: 10.18632/oncotarget.14143. Oncotarget. 2017. PMID: 28030794 Free PMC article.
-
A new perspective on diagnostic strategies concerning the potential of saliva-based miRNA signatures in oral cancer.Diagn Pathol. 2024 Nov 16;19(1):147. doi: 10.1186/s13000-024-01575-1. Diagn Pathol. 2024. PMID: 39548527 Free PMC article. Review.
-
Predictive Prognostic Value of Tissue-Based MicroRNA Expression in Oral Squamous Cell Carcinoma: A Systematic Review and Meta-analysis.J Dent Res. 2018 Jul;97(7):759-766. doi: 10.1177/0022034518762090. Epub 2018 Mar 13. J Dent Res. 2018. PMID: 29533734
Cited by
-
Identification of Prognosis Associated microRNAs in HNSCC Subtypes Based on TCGA Dataset.Medicina (Kaunas). 2020 Oct 13;56(10):535. doi: 10.3390/medicina56100535. Medicina (Kaunas). 2020. PMID: 33066067 Free PMC article.
-
MicroRNAs as Modulators of Oral Tumorigenesis-A Focused Review.Int J Mol Sci. 2021 Mar 4;22(5):2561. doi: 10.3390/ijms22052561. Int J Mol Sci. 2021. PMID: 33806361 Free PMC article. Review.
-
[Prognostic value of plasma miR-1290 expression in patients with oral squamous cell carcinoma].Hua Xi Kou Qiang Yi Xue Za Zhi. 2020 Aug 1;38(4):371-375. doi: 10.7518/hxkq.2020.04.003. Hua Xi Kou Qiang Yi Xue Za Zhi. 2020. PMID: 32865353 Free PMC article. Chinese.
-
Salivary Micro-RNA and Oral Squamous Cell Carcinoma: A Systematic Review.J Pers Med. 2021 Feb 4;11(2):101. doi: 10.3390/jpm11020101. J Pers Med. 2021. PMID: 33557138 Free PMC article. Review.
-
Exosomes in oral squamous cell carcinoma: functions, challenges, and potential applications.Front Oncol. 2025 Jan 16;14:1502283. doi: 10.3389/fonc.2024.1502283. eCollection 2024. Front Oncol. 2025. PMID: 39886659 Free PMC article. Review.
References
-
- Malik UU, Zarina S, Pennington SR. Oral squamous cell carcinoma: Key clinical questions, biomarker discovery, and the role of proteomics. Arch Oral Biol 2016;63:53–65. - PubMed
-
- Park EJ, Shimaoka M, Kiyono H. MicroRNA-mediated dynamic control of mucosal immunity. Int Immunol 2017;29:157–63. - PubMed
-
- Perez-Sayans M, Pilar GD, Barros-Angueira F, et al. Current trends in miRNAs and their relationship with oral squamous cell carcinoma. J Oral Pathol Med 2012;41:433–43. - PubMed
-
- Momen-Heravi F, Bala S. miRNA regulation of innate immunity. J Leukoc Biol 2018;Apr 14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

