A theoretical analysis of single molecule protein sequencing via weak binding spectra
- PMID: 30921350
- PMCID: PMC6438480
- DOI: 10.1371/journal.pone.0212868
A theoretical analysis of single molecule protein sequencing via weak binding spectra
Abstract
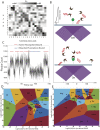
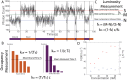
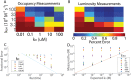
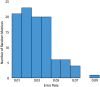
We propose and theoretically study an approach to massively parallel single molecule peptide sequencing, based on single molecule measurement of the kinetics of probe binding (Havranek, et al., 2013) to the N-termini of immobilized peptides. Unlike previous proposals, this method is robust to both weak and non-specific probe-target affinities, which we demonstrate by applying the method to a range of randomized affinity matrices consisting of relatively low-quality binders. This suggests a novel principle for proteomic measurement whereby highly non-optimized sets of low-affinity binders could be applicable for protein sequencing, thus shifting the burden of amino acid identification from biomolecular design to readout. Measurement of probe occupancy times, or of time-averaged fluorescence, should allow high-accuracy determination of N-terminal amino acid identity for realistic probe sets. The time-averaged fluorescence method scales well to weakly-binding probes with dissociation constants of tens or hundreds of micromolar, and bypasses photobleaching limitations associated with other fluorescence-based approaches to protein sequencing. We argue that this method could lead to an approach with single amino acid resolution and the ability to distinguish many canonical and modified amino acids, even using highly non-optimized probe sets. This readout method should expand the design space for single molecule peptide sequencing by removing constraints on the properties of the fluorescent binding probes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Real-time dynamic single-molecule protein sequencing on an integrated semiconductor device.Science. 2022 Oct 14;378(6616):186-192. doi: 10.1126/science.abo7651. Epub 2022 Oct 13. Science. 2022. PMID: 36227977
-
Leveraging nature's biomolecular designs in next-generation protein sequencing reagent development.Appl Microbiol Biotechnol. 2020 Sep;104(17):7261-7271. doi: 10.1007/s00253-020-10745-2. Epub 2020 Jul 2. Appl Microbiol Biotechnol. 2020. PMID: 32617618 Review.
-
A set of robust fluorescent peptide probes for quantification of Cu(ii) binding affinities in the micromolar to femtomolar range.Metallomics. 2015 Mar;7(3):567-78. doi: 10.1039/c4mt00301b. Metallomics. 2015. PMID: 25715324
-
An Approach for Peptide Identification by De Novo Sequencing of Mixture Spectra.IEEE/ACM Trans Comput Biol Bioinform. 2017 Mar-Apr;14(2):326-336. doi: 10.1109/TCBB.2015.2407401. IEEE/ACM Trans Comput Biol Bioinform. 2017. PMID: 28368810
-
Fluorescent Biosensors Based on Single-Molecule Counting.Acc Chem Res. 2016 Sep 20;49(9):1722-30. doi: 10.1021/acs.accounts.6b00237. Epub 2016 Sep 1. Acc Chem Res. 2016. PMID: 27583695 Review.
Cited by
-
Strategies for Development of a Next-Generation Protein Sequencing Platform.Trends Biochem Sci. 2020 Jan;45(1):76-89. doi: 10.1016/j.tibs.2019.09.005. Epub 2019 Oct 30. Trends Biochem Sci. 2020. PMID: 31676211 Free PMC article. Review.
-
Single-molecule fluorescence methods for protein biomarker analysis.Anal Bioanal Chem. 2023 Jul;415(18):3655-3669. doi: 10.1007/s00216-022-04502-9. Epub 2023 Jan 7. Anal Bioanal Chem. 2023. PMID: 36609860 Review.
-
Protein Sequencing, One Molecule at a Time.Annu Rev Biophys. 2022 May 9;51:181-200. doi: 10.1146/annurev-biophys-102121-103615. Epub 2022 Jan 5. Annu Rev Biophys. 2022. PMID: 34985940 Free PMC article. Review.
-
Computational assessment of the feasibility of protonation-based protein sequencing.PLoS One. 2020 Sep 11;15(9):e0238625. doi: 10.1371/journal.pone.0238625. eCollection 2020. PLoS One. 2020. PMID: 32915813 Free PMC article.
-
The emerging landscape of single-molecule protein sequencing technologies.Nat Methods. 2021 Jun;18(6):604-617. doi: 10.1038/s41592-021-01143-1. Epub 2021 Jun 7. Nat Methods. 2021. PMID: 34099939 Free PMC article. Review.
References
-
- Havranek JJ, Borgo B, inventors; Washington University in St Louis, assignee. Molecules and methods for iterative polypeptide analysis and processing. US20140273004A1; 2013. Available from: https://patents.google.com/patent/US20140273004A1/en.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

