Single-cell chromatin immunocleavage sequencing (scChIC-seq) to profile histone modification
- PMID: 30923384
- PMCID: PMC7187538
- DOI: 10.1038/s41592-019-0361-7
Single-cell chromatin immunocleavage sequencing (scChIC-seq) to profile histone modification
Abstract
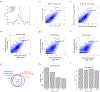
Our method for analyzing histone modifications, scChIC-seq (single-cell chromatin immunocleavage sequencing), involves targeting of the micrococcal nuclease (MNase) to a histone mark of choice by tethering to a specific antibody. Cleaved target sites are then selectively PCR amplified. We show that scChIC-seq reliably detects H3K4me3 and H3K27me3 target sites in single human white blood cells. The resulting data are used for clustering of blood cell types.
Figures






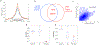


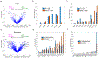


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

