Identification and Mapping of a 2,009-bp DNA Deletion in SERPING1 of a Hereditary Angioedema Patient
- PMID: 30923640
- PMCID: PMC6409050
- DOI: 10.1155/2019/7052062
Identification and Mapping of a 2,009-bp DNA Deletion in SERPING1 of a Hereditary Angioedema Patient
Abstract
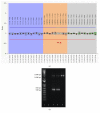
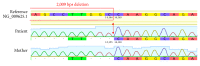
We report a heterozygous, 2,009 base pairs (bps) genomic DNA deletion within the SERPING1 gene that has not previously been reported in a case of type I hereditary angioedema (HAE). The patient is a 28-year-old Han Chinese female living in Hong Kong who has suffered from recurrent angioedema since adolescence, with increasing attack frequency as she entered adulthood; in the past, episodes occurred annually, but now occur every two to three months. The affected areas are not itchy and include common sites such as the left and right forearms, but without throat involvement. The patient also experiences epigastric pain. The patient's mother suffers from similar symptoms. A mutation in the serine protease inhibitor, clade G, member 1 (SERPING1) gene is associated with HAE. Patients with HAE type I commonly carry either a small deletion within SERPING1 or a truncated transcript. We performed a multiplex ligation-dependent probe amplification (MLPA) assay on our indexed patient. Our result suggests a 2,009 bps deletion spanning across exons 5 and 6 within SERPING1. Although earlier literature has described other large DNA deletions encasing exons 5 and 6 in SERPING1, these DNA rearrangements were larger in size between 4 and 6 kbps, and the breakpoint locations were generally not determined due to technical constraints (Pappalardo et al., 2000; Duponchel et al., 2001; Roche et al., 2005; Loules et al., 2018; and Göβwein et al., 2008). Our report describes mapping of this 2,009 bps in SERPING1. Using a combination of molecular techniques, we were able to confirm and locate this large heterozygous genomic DNA deletion that includes both exons 5 and 6 of SERPING1.
Figures

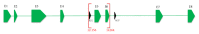
References
-
- Agostoni A., Aygören-Pürsün E., Binkley K. E., Blanch A., Bork K., Bouillet L., et al. Hereditary and acquired angioedema: problems and progress: proceedings of the third C1 esterase inhibitor deficiency workshop and beyond. The Journal of Allergy and Clinical Immunology. 2004;114(3):S51–S131. doi: 10.1016/j.jaci.2004.06.047. - DOI - PMC - PubMed
-
- Maurer M., Magerl M., Ansotegui I., Aygören-Pürsün E., Betschel S., Bork K., et al. The international WAO/EAACI guideline for the management of hereditary angioedema—The 2017 revision and update. Allergy: European Journal of Allergy and Clinical Immunology. 2018;73(8):1575–1596. doi: 10.1111/all.13384. - DOI - PubMed
-
- Bygum A., Fagerberg C. R., Ponard D., Monnier N., Lunardi J., Drouet C. Mutational spectrum and phenotypes in Danish families with hereditary angioedema because of C1 inhibitor deficiency. Allergy: European Journal of Allergy and Clinical Immunology. 2011;66(1):76–84. doi: 10.1111/j.1398-9995.2010.02456.x. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

