The Impact of Natural Selection on Short Insertion and Deletion Variation in the Great Tit Genome
- PMID: 30924871
- PMCID: PMC6543879
- DOI: 10.1093/gbe/evz068
The Impact of Natural Selection on Short Insertion and Deletion Variation in the Great Tit Genome
Abstract
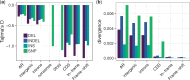
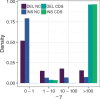
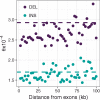
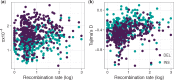
Insertions and deletions (INDELs) remain understudied, despite being the most common form of genetic variation after single nucleotide polymorphisms. This stems partly from the challenge of correctly identifying the ancestral state of an INDEL and thus identifying it as an insertion or a deletion. Erroneously assigned ancestral states can skew the site frequency spectrum, leading to artificial signals of selection. Consequently, the selective pressures acting on INDELs are, at present, poorly resolved. To tackle this issue, we have recently published a maximum likelihood approach to estimate the mutation rate and the distribution of fitness effects for INDELs. Our approach estimates and controls for the rate of ancestral state misidentification, overcoming issues plaguing previous INDEL studies. Here, we apply the method to INDEL polymorphism data from ten high coverage (∼44×) European great tit (Parus major) genomes. We demonstrate that coding INDELs are under strong purifying selection with a small proportion making it into the population (∼4%). However, among fixed coding INDELs, 71% of insertions and 86% of deletions are fixed by positive selection. In noncoding regions, we estimate ∼80% of insertions and ∼52% of deletions are effectively neutral, the remainder show signatures of purifying selection. Additionally, we see evidence of linked selection reducing INDEL diversity below background levels, both in proximity to exons and in areas of low recombination.
Keywords: adaptive mutation; deletions; distribution of fitness effects; insertions; linked selection.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
Similar articles
-
New Methods for Inferring the Distribution of Fitness Effects for INDELs and SNPs.Mol Biol Evol. 2018 Jun 1;35(6):1536-1546. doi: 10.1093/molbev/msy054. Mol Biol Evol. 2018. PMID: 29635416 Free PMC article.
-
Strong mutational bias toward deletions in the Drosophila melanogaster genome is compensated by selection.Genome Biol Evol. 2013;5(3):514-24. doi: 10.1093/gbe/evt021. Genome Biol Evol. 2013. PMID: 23395983 Free PMC article.
-
Strong heterogeneity in mutation rate causes misleading hallmarks of natural selection on indel mutations in the human genome.Mol Biol Evol. 2014 Jan;31(1):23-36. doi: 10.1093/molbev/mst185. Epub 2013 Oct 9. Mol Biol Evol. 2014. PMID: 24113537 Free PMC article.
-
The repeatability of genome-wide mutation rate and spectrum estimates.Curr Genet. 2016 Aug;62(3):507-12. doi: 10.1007/s00294-016-0573-7. Epub 2016 Feb 26. Curr Genet. 2016. PMID: 26919990 Free PMC article. Review.
-
The association of insertions/deletions (INDELs) and variable number tandem repeats (VNTRs) with obesity and its related traits and complications.J Physiol Anthropol. 2017 Jun 14;36(1):25. doi: 10.1186/s40101-017-0142-x. J Physiol Anthropol. 2017. PMID: 28615046 Free PMC article. Review.
Cited by
-
Channel nuclear pore protein 54 directs sexual differentiation and neuronal wiring of female reproductive behaviors in Drosophila.BMC Biol. 2021 Oct 20;19(1):226. doi: 10.1186/s12915-021-01154-6. BMC Biol. 2021. PMID: 34666772 Free PMC article.
-
Comparative population pangenomes reveal unexpected complexity and fitness effects of structural variants.bioRxiv [Preprint]. 2025 Feb 13:2025.02.11.637762. doi: 10.1101/2025.02.11.637762. bioRxiv. 2025. PMID: 39990470 Free PMC article. Preprint.
-
Purifying selection on noncoding deletions of human regulatory loci detected using their cellular pleiotropy.Genome Res. 2021 Jun;31(6):935-946. doi: 10.1101/gr.275263.121. Epub 2021 May 7. Genome Res. 2021. PMID: 33963077 Free PMC article.
-
A Study of Faster-Z Evolution in the Great Tit (Parus major).Genome Biol Evol. 2020 Mar 1;12(3):210-222. doi: 10.1093/gbe/evaa044. Genome Biol Evol. 2020. PMID: 32119100 Free PMC article.
-
Insertions and Deletions: Computational Methods, Evolutionary Dynamics, and Biological Applications.Mol Biol Evol. 2024 Sep 4;41(9):msae177. doi: 10.1093/molbev/msae177. Mol Biol Evol. 2024. PMID: 39172750 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

