Adaptive Strategies of the Candidate Probiotic E. coli Nissle in the Mammalian Gut
- PMID: 30926240
- PMCID: PMC6487504
- DOI: 10.1016/j.chom.2019.02.005
Adaptive Strategies of the Candidate Probiotic E. coli Nissle in the Mammalian Gut
Abstract
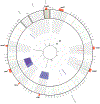
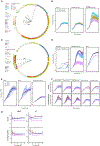
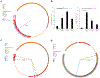
Probiotics are living microorganisms that are increasingly used as gastrointestinal therapeutics by virtue of their innate or engineered genetic function. Unlike abiotic therapeutics, probiotics can replicate in their intended site, subjecting their genomes and therapeutic properties to natural selection. We exposed the candidate probiotic E. coli Nissle (EcN) to the mouse gastrointestinal tract over several weeks, systematically altering the diet and background microbiota complexity. In-transit EcN accumulates genetic mutations that modulate carbohydrate utilization, stress response, and adhesion to gain competitive fitness, while previous exposure to antibiotics reveals an acquisition of resistance. We then leveraged these insights to generate an EcN strain that shows therapeutic efficacy in a mouse model of phenylketonuria and found that it was genetically stable over 1 week, thereby validating EcN's utility as a chassis for engineering. Collectively, we demonstrate a generalizable pipeline that can be applied to other probiotics to better understand their safety and engineering potential.
Keywords: E. coli; engineering; evolution; microbiome; phenylketonuria; probiotic.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests
The authors declare no competing interests. The following authors have the current affiliations: AJG: VL55; MKG: Flagship Pioneering; DP: Eli Lilly & Company. The authors assert that VL55, Flagship Pioneering, and Eli Lilly & Company were not involved in and did not influence this research.
Figures




Comment in
-
Can the Microbiome Deliver? A Proof-of-Concept Engineered E. coli PKU Therapeutic.Cell Host Microbe. 2019 Apr 10;25(4):473-474. doi: 10.1016/j.chom.2019.03.015. Cell Host Microbe. 2019. PMID: 30974078
References
-
- Anderson C (2006. ). Anderson promoter collection. http://parts.igem.org/Promoters/Catalog/Anderson
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

