Hepatic Lysosomal iNOS Activity Impairs Autophagy in Obesity
- PMID: 30926581
- PMCID: PMC6522853
- DOI: 10.1016/j.jcmgh.2019.03.005
Hepatic Lysosomal iNOS Activity Impairs Autophagy in Obesity
Abstract
Background & aims: The lysosome is an acidic organelle that is important for maintaining cellular and metabolic homeostasis in hepatocytes. Lysosomal dysfunction and chronic inflammation coexist, and both contribute to obesity-associated hepatic insulin resistance. However, in the context of obesity, the interplay between inflammatory signals and hepatic lysosomal function remains largely unknown. Inducible nitric oxide synthase (iNOS) is a hallmark for inflammation, and is activated in obesity. The aim of this study is to understand the molecular link between iNOS-mediated lysosomal nitric oxide (NO) production, hepatic lysosomal function, and autophagy in the context of obesity-associated insulin resistance.
Methods: The role of iNOS in hepatic autophagy, as related to insulin and glucose homeostasis were studied in mice with diet-induced obesity (DIO). The effects and mechanisms of iNOS-mediated lysosomal NO production on lysosomal function and hepatic autophagy were studied in primary hepatocytes as well as in a mouse model of DIO.
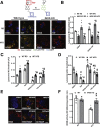
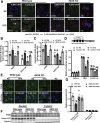
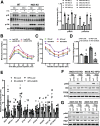
Results: We demonstrate that obesity promotes iNOS localization to the lysosome and decreases levels of lysosomal arginine, resulting in an accumulation of NO in hepatic lysosomes. This lysosomal NO production is attenuated by treatment with a NO scavenger, while co-overexpression of mTOR and a lysosomal arginine transporter (SLC38A9) enhances lysosomal NO production and suppresses autophagy. In addition, we show that deletion of iNOS ameliorates lysosomal nitrosative stress in the livers of DIO mice, promotes lysosomal biogenesis by activating transcription factor EB (TFEB), and enhances lysosomal function and autophagy. Lastly, deletion of iNOS in mice with DIO improves hepatic insulin sensitivity, which is diminished by suppression of TFEB or autophagy related 7 (Atg7).
Conclusions: Our studies suggest that lysosomal iNOS-mediated NO signaling disrupts hepatic lysosomal function, contributing to obesity-associated defective hepatic autophagy and insulin resistance.
Keywords: Autophagy; Inducible Nitric Oxide Synthase; Insulin Resistance; Lysosome; Nitric Oxide.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





Comment in
-
NO to Lysosomes: A Signal for Insulin Resistance in Obesity.Cell Mol Gastroenterol Hepatol. 2019;8(1):153-154. doi: 10.1016/j.jcmgh.2019.04.006. Epub 2019 May 15. Cell Mol Gastroenterol Hepatol. 2019. PMID: 31102588 Free PMC article. No abstract available.
References
-
- Kuntz E., Kuntz H.D. Springer; Berlin: 2002. Hepatology, Principles, and Practice: History, Morphology, Biochemistry, Diagnostics, Clinic, Therapy.
-
- Codogno P., Meijer A.J. Autophagy in the liver. J Hepatol. 2013;59:389–391. - PubMed
-
- Doria A., Gatto M., Punzi L. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

