Active-site differences between substrate-free and ritonavir-bound cytochrome P450 (CYP) 3A5 reveal plasticity differences between CYP3A5 and CYP3A4
- PMID: 30926609
- PMCID: PMC6527174
- DOI: 10.1074/jbc.RA119.007928
Active-site differences between substrate-free and ritonavir-bound cytochrome P450 (CYP) 3A5 reveal plasticity differences between CYP3A5 and CYP3A4
Abstract
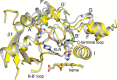
Cytochrome P450 (CYP) 3A4 is a major contributor to hepatic drug and xenobiotic metabolism in human adults. The related enzyme CYP3A5 is also expressed in adult liver and has broader age and tissue distributions. However, CYP3A5 expression is low in most Caucasians because of the prevalence of an allele that leads to an incorrectly spliced mRNA and premature termination of translation. When expressed, CYP3A5 expands metabolic capabilities and can augment CYP3A4-mediated drug metabolism, thereby reducing drug efficacy and potentially requiring dose adjustments. The extensive role of CYP3A4 in drug metabolism reflects in part the plasticity of the substrate-free enzyme to enlarge its active site and accommodate very large substrates. We have previously shown that the structure of the CYP3A5-ritonavir complex differs substantially from that of the CYP3A4-ritonavir complex. To better understand whether these differences are conserved in other CYP3A5 structures and how they relate to differential plasticity, we determined the X-ray crystallographic structure of the CYP3A5 substrate-free complex to 2.20 Å resolution. We observed that this structure exhibits a much larger active site than substrate-free CYP3A4 and displays an open substrate access channel. This reflected in part a lower trajectory of the helix F-F' connector in CYP3A4 and more extensive π-CH interactions between phenylalanine residues forming the roof of the active-site cavity than in CYP3A5. Comparison with the CYP3A5-ritonavir complex confirmed conserved CYP3A5 structural features and indicated differences in plasticity between CYP3A4 and CYP3A5 that favor alternative ritonavir conformations.
Keywords: CYP3A4; CYP3A5; crystal structure; cytochrome P450; drug metabolism; protein conformation; protein plasticity; protein structure.
© 2019 Hsu and Johnson.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Aoyama T., Yamano S., Waxman D. J., Lapenson D. P., Meyer U. A., Fischer V., Tyndale R., Inaba T., Kalow W., and Gelboin H. V. (1989) Cytochrome P-450 hPCN3, a novel cytochrome P-450 IIIA gene product that is differentially expressed in adult human liver. J. Biol. Chem. 264, 10388–10395 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

