Precise small-molecule cleavage of an r(CUG) repeat expansion in a myotonic dystrophy mouse model
- PMID: 30926669
- PMCID: PMC6475439
- DOI: 10.1073/pnas.1901484116
Precise small-molecule cleavage of an r(CUG) repeat expansion in a myotonic dystrophy mouse model
Abstract
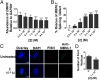
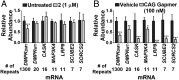
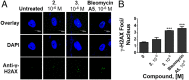
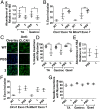
Myotonic dystrophy type 1 (DM1) is an incurable neuromuscular disorder caused by an expanded CTG repeat that is transcribed into r(CUG)exp The RNA repeat expansion sequesters regulatory proteins such as Muscleblind-like protein 1 (MBNL1), which causes pre-mRNA splicing defects. The disease-causing r(CUG)exp has been targeted by antisense oligonucleotides, CRISPR-based approaches, and RNA-targeting small molecules. Herein, we describe a designer small molecule, Cugamycin, that recognizes the structure of r(CUG)exp and cleaves it in both DM1 patient-derived myotubes and a DM1 mouse model, leaving short repeats of r(CUG) untouched. In contrast, oligonucleotides that recognize r(CUG) sequence rather than structure cleave both long and short r(CUG)-containing transcripts. Transcriptomic, histological, and phenotypic studies demonstrate that Cugamycin broadly and specifically relieves DM1-associated defects in vivo without detectable off-targets. Thus, small molecules that bind and cleave RNA have utility as lead chemical probes and medicines and can selectively target disease-causing RNA structures to broadly improve defects in preclinical animal models.
Keywords: RNA; RNA splicing; chemical biology; genetic disease; nucleic acids.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
Conflict of interest statement: M.D.D. and E.T.W. are consultants for Expansion Therapeutics. S.G.R. is a current employee of Expansion Therapeutics.
Figures


References
-
- Schlünzen F, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. - PubMed
-
- Carter AP, et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

