Genomic and transcriptomic changes complement each other in the pathogenesis of sporadic Burkitt lymphoma
- PMID: 30926794
- PMCID: PMC6440956
- DOI: 10.1038/s41467-019-08578-3
Genomic and transcriptomic changes complement each other in the pathogenesis of sporadic Burkitt lymphoma
Abstract
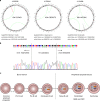
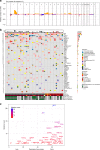
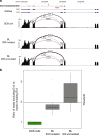
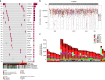
Burkitt lymphoma (BL) is the most common B-cell lymphoma in children. Within the International Cancer Genome Consortium (ICGC), we performed whole genome and transcriptome sequencing of 39 sporadic BL. Here, we unravel interaction of structural, mutational, and transcriptional changes, which contribute to MYC oncogene dysregulation together with the pathognomonic IG-MYC translocation. Moreover, by mapping IGH translocation breakpoints, we provide evidence that the precursor of at least a subset of BL is a B-cell poised to express IGHA. We describe the landscape of mutations, structural variants, and mutational processes, and identified a series of driver genes in the pathogenesis of BL, which can be targeted by various mechanisms, including IG-non MYC translocations, germline and somatic mutations, fusion transcripts, and alternative splicing.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Swerdlow, S. et al. in WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues (ed Swerdlow, S. et al.) (International Agency for Research on Cancer, Lyon, France, 2017).
-
- Burkhardt B, et al. Non-Hodgkin's lymphoma in adolescents: experiences in 378 adolescent NHL patients treated according to pediatric NHL-BFM protocols. Leukemia. 2011;25:153–160. - PubMed
-
- Miles RR, Arnold S, Cairo MS. Risk factors and treatment of childhood and adolescent Burkitt lymphoma/leukaemia. Br. J. Haematol. 2012;156:730–743. - PubMed
-
- Burkitt DP. Classics in oncology. A sarcoma involving the jaws in African children. CA Cancer J. Clin. 1972;22:345–355. - PubMed
-
- Gabarre J, et al. Human immunodeficiency virus-related lymphoma: relation between clinical features and histologic subtypes. Am. J. Med. 2001;111:704–711. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

