Opportunities and challenges for transcriptome-wide association studies
- PMID: 30926968
- PMCID: PMC6777347
- DOI: 10.1038/s41588-019-0385-z
Opportunities and challenges for transcriptome-wide association studies
Abstract
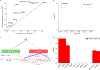
Transcriptome-wide association studies (TWAS) integrate genome-wide association studies (GWAS) and gene expression datasets to identify gene-trait associations. In this Perspective, we explore properties of TWAS as a potential approach to prioritize causal genes at GWAS loci, by using simulations and case studies of literature-curated candidate causal genes for schizophrenia, low-density-lipoprotein cholesterol and Crohn's disease. We explore risk loci where TWAS accurately prioritizes the likely causal gene as well as loci where TWAS prioritizes multiple genes, some likely to be non-causal, owing to sharing of expression quantitative trait loci (eQTL). TWAS is especially prone to spurious prioritization with expression data from non-trait-related tissues or cell types, owing to substantial cross-cell-type variation in expression levels and eQTL strengths. Nonetheless, TWAS prioritizes candidate causal genes more accurately than simple baselines. We suggest best practices for causal-gene prioritization with TWAS and discuss future opportunities for improvement. Our results showcase the strengths and limitations of using eQTL datasets to determine causal genes at GWAS loci.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures

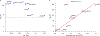
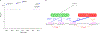
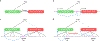
References
-
- Zhu Z et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet 48, 481–487 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 5U01HG009080 /NH/NIH HHS/United States
- U01 HG009431/HG/NHGRI NIH HHS/United States
- R01HG009120 /NH/NIH HHS/United States
- R01 MH107666/MH/NIMH NIH HHS/United States
- R01 HG006399/HG/NHGRI NIH HHS/United States
- 1U24HG008956 /NH/NIH HHS/United States
- R21TR001739 /NH/NIH HHS/United States
- U01HG009431 /NH/NIH HHS/United States
- U01 HG009080/HG/NHGRI NIH HHS/United States
- R01 MH101820/MH/NIMH NIH HHS/United States
- R01MH101820 /NH/NIH HHS/United States
- P30 DK116074/DK/NIDDK NIH HHS/United States
- R21 TR001739/TR/NCATS NIH HHS/United States
- U24 HG008956/HG/NHGRI NIH HHS/United States
- P30 DK020595/DK/NIDDK NIH HHS/United States
- R01 HL125863/HL/NHLBI NIH HHS/United States
- 1DP2OD022870 /NH/NIH HHS/United States
- R01MH107666/NH/NIH HHS/United States
- P30DK20595 /NH/NIH HHS/United States
- R01HL125863 /NH/NIH HHS/United States
- R01 HG010140/HG/NHGRI NIH HHS/United States
- R01 MH115676/MH/NIMH NIH HHS/United States
- R01MH115676 /NH/NIH HHS/United States
- DP2 GM123485/GM/NIGMS NIH HHS/United States
- R01 HG009120/HG/NHGRI NIH HHS/United States
- R01HG010140 /NH/NIH HHS/United States

