Genomic subtyping and therapeutic targeting of acute erythroleukemia
- PMID: 30926971
- PMCID: PMC6828160
- DOI: 10.1038/s41588-019-0375-1
Genomic subtyping and therapeutic targeting of acute erythroleukemia
Abstract
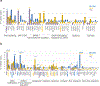
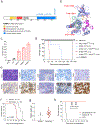
Acute erythroid leukemia (AEL) is a high-risk leukemia of poorly understood genetic basis, with controversy regarding diagnosis in the spectrum of myelodysplasia and myeloid leukemia. We compared genomic features of 159 childhood and adult AEL cases with non-AEL myeloid disorders and defined five age-related subgroups with distinct transcriptional profiles: adult, TP53 mutated; NPM1 mutated; KMT2A mutated/rearranged; adult, DDX41 mutated; and pediatric, NUP98 rearranged. Genomic features influenced outcome, with NPM1 mutations and HOXB9 overexpression being associated with a favorable prognosis and TP53, FLT3 or RB1 alterations associated with poor survival. Targetable signaling mutations were present in 45% of cases and included recurrent mutations of ALK and NTRK1, the latter of which drives erythroid leukemogenesis sensitive to TRK inhibition. This genomic landscape of AEL provides the framework for accurate diagnosis and risk stratification of this disease, and the rationale for testing targeted therapies in this high-risk leukemia.
Figures



References
-
- Cervera N et al. Molecular characterization of acute erythroid leukemia (M6-AML) using targeted next-generation sequencing. Leukemia 30, 966–70 (2016). - PubMed
-
- Grossmann V et al. Acute erythroid leukemia (AEL) can be separated into distinct prognostic subsets based on cytogenetic and molecular genetic characteristics. Leukemia 27, 1940–3 (2013). - PubMed
-
- Ping N et al. Exome sequencing identifies highly recurrent somatic GATA2 and CEBPA mutations in acute erythroid leukemia. Leukemia 31, 195–202 (2017). - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

