4E-BP1 and 4E-BP2 double knockout mice are protected from aging-associated sarcopenia
- PMID: 30927336
- PMCID: PMC6596930
- DOI: 10.1002/jcsm.12412
4E-BP1 and 4E-BP2 double knockout mice are protected from aging-associated sarcopenia
Abstract
Background: Sarcopenia is the loss of muscle mass/function that occurs during the aging process. The links between mechanistic target of rapamycin (mTOR) activity and muscle development are largely documented, but the role of its downstream targets in the development of sarcopenia is poorly understood. Eukaryotic initiation factor 4E-binding proteins (4E-BPs) are targets of mTOR that repress mRNA translation initiation and are involved in the control of several physiological processes. However, their role in skeletal muscle is still poorly understood. The goal of this study was to assess how loss of 4E-BP1 and 4E-BP2 expression impacts skeletal muscle function and homeostasis in aged mice and to characterize the associated metabolic changes by metabolomic and lipidomic profiling.
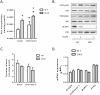
Methods: Twenty-four-month-old wild-type and whole body 4E-BP1/4E-BP2 double knockout (DKO) mice were used to measure muscle mass and function. Protein homeostasis was measured ex vivo in extensor digitorum longus by incorporation of l-[U-14 C]phenylalanine, and metabolomic and lipidomic profiling of skeletal muscle was performed by Metabolon, Inc.
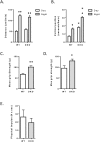
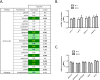
Results: The 4E-BP1/2 DKO mice exhibited an increase in muscle mass that was associated with increased grip strength (P < 0.05). Protein synthesis was higher under both basal (+102%, P < 0.05) and stimulated conditions (+65%, P < 0.05) in DKO skeletal muscle. Metabolomic and complex lipid analysis of skeletal muscle revealed robust differences pertaining to amino acid homeostasis, carbohydrate abundance, and certain aspects of lipid metabolism. In particular, levels of most free amino acids were lower within the 4E-BP1/2 DKO muscle. Interestingly, although glucose levels were unchanged, differences were observed in the isobaric compound maltitol/lactitol (33-fold increase, P < 0.01) and in several additional carbohydrate compounds. 4E-BP1/2 depletion also resulted in accumulation of medium-chain acylcarnitines and a 20% lower C2/C0 acylcarnitine ratio (P < 0.01) indicative of reduced β-oxidation.
Conclusions: Taken together, these findings demonstrate that deletion of 4E-BPs is associated with perturbed energy metabolism in skeletal muscle and could have beneficial effects on skeletal muscle mass and function in aging mice. They also identify 4E-BPs as potential targets for the treatment of sarcopenia.
Keywords: Anabolism; Protein synthesis; Proteolysis; Skeletal muscle; mTOR.
© 2019 The Authors. Journal of Cachexia, Sarcopenia and Muscle published by John Wiley & Sons Ltd on behalf of the Society on Sarcopenia, Cachexia and Wasting Disorders.
Conflict of interest statement
The authors declare they have no conflict of interest.
Figures


References
-
- Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC Jr, et al. Insulin‐dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′‐cap function. Nature 1994;371:762–767. - PubMed
-
- Poulin F, Gingras AC, Olsen H, Chevalier S, Sonenberg N. 4E‐BP3, a new member of the eukaryotic initiation factor 4E‐binding protein family. J Biol Chem 1998;273:14002–14007. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

