Immune Evasion by Staphylococcus aureus
- PMID: 30927347
- PMCID: PMC11590434
- DOI: 10.1128/microbiolspec.GPP3-0061-2019
Immune Evasion by Staphylococcus aureus
Abstract
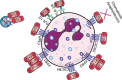
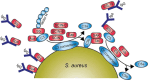
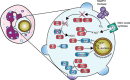
Staphylococcus aureus has become a serious threat to human health. In addition to having increased antibiotic resistance, the bacterium is a master at adapting to its host by evading almost every facet of the immune system, the so-called immune evasion proteins. Many of these immune evasion proteins target neutrophils, the most important immune cells in clearing S. aureus infections. The neutrophil attacks pathogens via a plethora of strategies. Therefore, it is no surprise that S. aureus has evolved numerous immune evasion strategies at almost every level imaginable. In this review we discuss step by step the aspects of neutrophil-mediated killing of S. aureus, such as neutrophil activation, migration to the site of infection, bacterial opsonization, phagocytosis, and subsequent neutrophil-mediated killing. After each section we discuss how S. aureus evasion molecules are able to resist the neutrophil attack of these different steps. To date, around 40 immune evasion molecules of S. aureus are known, but its repertoire is still expanding due to the discovery of new evasion proteins and the addition of new functions to already identified evasion proteins. Interestingly, because the different parts of neutrophil attack are redundant, the evasion molecules display redundant functions as well. Knowing how and with which proteins S. aureus is evading the immune system is important in understanding the pathophysiology of this pathogen. This knowledge is crucial for the development of therapeutic approaches that aim to clear staphylococcal infections.
Figures



References
-
- Gorwitz RJ, Kruszon-Moran D, McAllister SK, McQuillan G, McDougal LK, Fosheim GE, Jensen BJ, Killgore G, Tenover FC, Kuehnert MJ. 2008. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001-2004. J Infect Dis 197:1226–1234 10.1086/533494. [PubMed] - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

