MYC Recruits SPT5 to RNA Polymerase II to Promote Processive Transcription Elongation
- PMID: 30928206
- PMCID: PMC6527870
- DOI: 10.1016/j.molcel.2019.02.031
MYC Recruits SPT5 to RNA Polymerase II to Promote Processive Transcription Elongation
Abstract
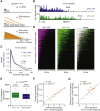
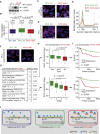
The MYC oncoprotein binds to promoter-proximal regions of virtually all transcribed genes and enhances RNA polymerase II (Pol II) function, but its precise mode of action is poorly understood. Using mass spectrometry of both MYC and Pol II complexes, we show here that MYC controls the assembly of Pol II with a small set of transcription elongation factors that includes SPT5, a subunit of the elongation factor DSIF. MYC directly binds SPT5, recruits SPT5 to promoters, and enables the CDK7-dependent transfer of SPT5 onto Pol II. Consistent with known functions of SPT5, MYC is required for fast and processive transcription elongation. Intriguingly, the high levels of MYC that are expressed in tumors sequester SPT5 into non-functional complexes, thereby decreasing the expression of growth-suppressive genes. Altogether, these results argue that MYC controls the productive assembly of processive Pol II elongation complexes and provide insight into how oncogenic levels of MYC permit uncontrolled cellular growth.
Keywords: MYC; RNA polymerase II; SPT5; SPT6; SUPT5H; directionality; elongation rate; processivity; transcription; tumorigenesis.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Bernecky C., Plitzko J.M., Cramer P. Structure of a transcribing RNA polymerase II-DSIF complex reveals a multidentate DNA-RNA clamp. Nat. Struct. Mol. Biol. 2017;24:809–815. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

