Systematic evaluation of the microRNAome through miR-CATCHv2.0 identifies positive and negative regulators of BRAF-X1 mRNA
- PMID: 30929607
- PMCID: PMC6546394
- DOI: 10.1080/15476286.2019.1600934
Systematic evaluation of the microRNAome through miR-CATCHv2.0 identifies positive and negative regulators of BRAF-X1 mRNA
Abstract
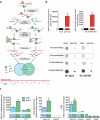
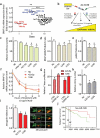
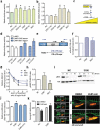
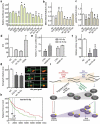
Here we present miR-CATCHv2.0, an implemented experimental method that allows the identification of the microRNA species directly bound to an RNA of interest. After cross-linking of microRNA::RNA::Ago2 complexes using formaldehyde, the RNA is fragmented using sonication and then subjected to affinity purification using two sets of biotinylated tiling probes (ODD and EVEN). Finally, enriched microRNA species are retrieved by means of small RNA sequencing coupled with an ad hoc analytical workflow. In BRAFV600E mutant A375 melanoma cells, miR-CATCHv2.0 allowed us to identify 20 microRNAs that target X1, the most abundant isoform of BRAF mRNA. These microRNAs fall into different functional classes, according to the effect that they exert (decrease/increase in BRAFV600E mRNA and protein levels) and to the mechanism they use to achieve it (destabilization/stabilization of X1 mRNA or decrease/increase in its translation). microRNA-induced variations in BRAFV600E protein levels are most of the times coupled to consistent variations in pMEK levels, in melanoma cell proliferation in vitro and in sensitivity to the BRAF inhibitor vemurafenib in a xenograft model in zebrafish. However, microRNAs exist that uncouple the degree of activation of the ERK pathway from the levels of BRAFV600E protein. Our study proposes miR-CATCHv2.0 as an effective tool for the identification of direct microRNA-target interactions and, by using such a tool, unveils the complexity of the post-transcriptional regulation to which BRAFV600E and the ERK pathway are subjected in melanoma cells.
Keywords: -X1 mRNA; Direct microRNA-target binding; ERK signalling; affinity purification; melanoma; transcript stability; transcript translation.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
