Genetic Architectures of Childhood- and Adult-Onset Asthma Are Partly Distinct
- PMID: 30929738
- PMCID: PMC6451732
- DOI: 10.1016/j.ajhg.2019.02.022
Genetic Architectures of Childhood- and Adult-Onset Asthma Are Partly Distinct
Abstract
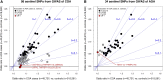
The extent to which genetic risk factors are shared between childhood-onset (COA) and adult-onset (AOA) asthma has not been estimated. On the basis of data from the UK Biobank study (n = 447,628), we found that the variance in disease liability explained by common variants is higher for COA (onset at ages between 0 and 19 years; h2g = 25.6%) than for AOA (onset at ages between 20 and 60 years; h2g = 10.6%). The genetic correlation (rg) between COA and AOA was 0.67. Variation in age of onset among COA-affected individuals had a low heritability (h2g = 5%), which we confirmed in independent studies and also among AOA-affected individuals. To identify subtype-specific genetic associations, we performed a genome-wide association study (GWAS) in the UK Biobank for COA (13,962 affected individuals) and a separate GWAS for AOA (26,582 affected individuals) by using a common set of 300,671 controls for both studies. We identified 123 independent associations for COA and 56 for AOA (37 overlapped); of these, 98 and 34, respectively, were reproducible in an independent study (n = 262,767). Collectively, 28 associations were not previously reported. For 96 COA-associated variants, including five variants that represent COA-specific risk factors, the risk allele was more common in COA- than in AOA-affected individuals. Conversely, we identified three variants that are stronger risk factors for AOA. Variants associated with obesity and smoking had a stronger contribution to the risk of AOA than to the risk of COA. Lastly, we identified 109 likely target genes of the associated variants, primarily on the basis of correlated expression quantitative trait loci (up to n = 31,684). GWAS informed by age of onset can identify subtype-specific risk variants, which can help us understand differences in pathophysiology between COA and AOA and so can be informative for drug development.
Keywords: GWAS; age; allergy; asthma; genetic; genome; heritability; onset; overlap; risk.
Copyright © 2019 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Tan D.J., Walters E.H., Perret J.L., Lodge C.J., Lowe A.J., Matheson M.C., Dharmage S.C. Age-of-asthma onset as a determinant of different asthma phenotypes in adults: A systematic review and meta-analysis of the literature. Expert Rev. Respir. Med. 2015;9:109–123. - PubMed
-
- Moore W.C., Meyers D.A., Wenzel S.E., Teague W.G., Li H., Li X., D’Agostino R., Jr., Castro M., Curran-Everett D., Fitzpatrick A.M., National Heart, Lung, and Blood Institute’s Severe Asthma Research Program Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am. J. Respir. Crit. Care Med. 2010;181:315–323. - PMC - PubMed
-
- Newby C., Heaney L.G., Menzies-Gow A., Niven R.M., Mansur A., Bucknall C., Chaudhuri R., Thompson J., Burton P., Brightling C., British Thoracic Society Severe Refractory Asthma Network Statistical cluster analysis of the British Thoracic Society Severe refractory Asthma Registry: Clinical outcomes and phenotype stability. PLoS ONE. 2014;9:e102987. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

