Transcriptional and Chemical Changes in Soybean Leaves in Response to Long-Term Aphid Colonization
- PMID: 30930925
- PMCID: PMC6424911
- DOI: 10.3389/fpls.2019.00310
Transcriptional and Chemical Changes in Soybean Leaves in Response to Long-Term Aphid Colonization
Abstract
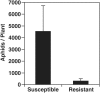
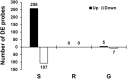
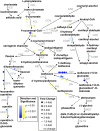
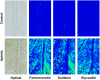
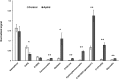
Soybean aphids (Aphis glycines Matsumura) are specialized insects that feed on soybean (Glycine max) phloem sap. Transcriptome analyses have shown that resistant soybean plants mount a fast response that limits aphid feeding and population growth. Conversely, defense responses in susceptible plants are slower and it is hypothesized that aphids block effective defenses in the compatible interaction. Unlike other pests, aphids can colonize plants for long periods of time; yet the effect on the plant transcriptome after long-term aphid feeding has not been analyzed for any plant-aphid interaction. We analyzed the susceptible and resistant (Rag1) transcriptome response to aphid feeding in soybean plants colonized by aphids (biotype 1) for 21 days. We found a reduced resistant response and a low level of aphid growth on Rag1 plants, while susceptible plants showed a strong response consistent with pattern-triggered immunity. GO-term analyses identified chitin regulation as one of the most overrepresented classes of genes, suggesting that chitin could be one of the hemipteran-associated molecular pattern that triggers this defense response. Transcriptome analyses also indicated the phenylpropanoid pathway, specifically isoflavonoid biosynthesis, was induced in susceptible plants in response to long-term aphid feeding. Metabolite analyses corroborated this finding. Aphid-treated susceptible plants accumulated daidzein, formononetin, and genistein, although glyceollins were present at low levels in these plants. Choice experiments indicated that daidzein may have a deterrent effect on aphid feeding. Mass spectrometry imaging showed these isoflavones accumulate likely in the mesophyll cells or epidermis and are absent from the vasculature, suggesting that isoflavones are part of a non-phloem defense response that can reduce aphid feeding. While it is likely that aphid can initially block defense responses in compatible interactions, it appears that susceptible soybean plants can eventually mount an effective defense in response to long-term soybean aphid colonization.
Keywords: Aphis glycines; feeding deterrence; isoflavones; plant–insect interaction; soybean defense.
Figures



References
-
- Akashi T., Sasaki K., Aoki T., Ayabe S., Yazaki K. (2009). Molecular cloning and characterization of a cDNA for pterocarpan 4-dimethylallyltransferase catalyzing the key prenylation step in the biosynthesis of glyceollin, a soybean phytoalexin. Plant Physiol. 149 683–693. 10.1104/pp.108.123679 - DOI - PMC - PubMed
-
- Alvarez A. E., Tjallingii W. F., Garzo E., Vleeshouwers V., Dicke M., Vosman B. (2006). Location of resistance factors in the leaves of potato and wild tuber-bearing Solanum species to the aphid Myzus persicae. Entomol. Exp. Et Appl. 121 145–157. 10.1111/j.1570-8703.2006.00464.x - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

