The survey and reference assisted assembly of the Octopus vulgaris genome
- PMID: 30931949
- PMCID: PMC6472339
- DOI: 10.1038/s41597-019-0017-6
The survey and reference assisted assembly of the Octopus vulgaris genome
Abstract
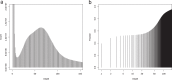
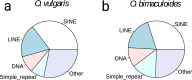
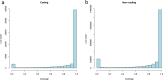
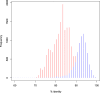
The common octopus, Octopus vulgaris, is an active marine predator known for the richness and plasticity of its behavioral repertoire, and remarkable learning and memory capabilities. Octopus and other coleoid cephalopods, cuttlefish and squid, possess the largest nervous system among invertebrates, both for cell counts and body to brain size. O. vulgaris has been at the center of a long-tradition of research into diverse aspects of its biology. To leverage research in this iconic species, we generated 270 Gb of genomic sequencing data, complementing those available for the only other sequenced congeneric octopus, Octopus bimaculoides. We show that both genomes are similar in size, but display different levels of heterozygosity and repeats. Our data give a first quantitative glimpse into the rate of coding and non-coding regions and support the view that hundreds of novel genes may have arisen independently despite the close phylogenetic distance. We furthermore describe a reference-guided assembly and an open genomic resource (CephRes-gdatabase), opening new avenues in the study of genomic novelties in cephalopods and their biology.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- De Luca D, Catanese G, Procaccini G, Fiorito G. An integration of historical records and genetic data to the assessment of global distribution and population structure in Octopus vulgaris. Front. Ecol. Evol. 2014;2:55.
-
- Amor MD, et al. Morphological assessment of the Octopus vulgaris species complex evaluated in light of molecular-based phylogenetic inferences. Zool. Scr. 2017;46:275–288. doi: 10.1111/zsc.12207. - DOI
-
- Wells, M. J. Octopus: physiology and behaviour of an advanced invertebrate. (Springer Science & Business Media, 1978).
-
- Marini, G., De Sio, F., Ponte, G. & Fiorito, G. In Learning and Memory: A Comprehensive Reference (Second Edition) Vol. Volume 1 - Learning Theory and Behavior (Menzel, Randolf - volume Editor) (ed John H. Byrne) 441–462 (Academic Press, Elsevier, 2017).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

