High-throughput, label-free, single-cell photoacoustic microscopy of intratumoral metabolic heterogeneity
- PMID: 30936431
- PMCID: PMC6544054
- DOI: 10.1038/s41551-019-0376-5
High-throughput, label-free, single-cell photoacoustic microscopy of intratumoral metabolic heterogeneity
Abstract
Intratumoral heterogeneity, which is manifested in almost all of the hallmarks of cancer, including the significantly altered metabolic profiles of cancer cells, represents a challenge to effective cancer therapy. High-throughput measurements of the metabolism of individual cancer cells would allow direct visualization and quantification of intratumoral metabolic heterogeneity, yet the throughputs of current measurement techniques are limited to about 120 cells per hour. Here, we show that single-cell photoacoustic microscopy can reach throughputs of approximately 12,000 cells per hour by trapping single cells with blood in an oxygen-diffusion-limited high-density microwell array and by using photoacoustic imaging to measure the haemoglobin oxygen change (that is, the oxygen consumption rate) in the microwells. We demonstrate the capability of this label-free technique by performing high-throughput single-cell oxygen-consumption-rate measurements of cultured cells and by imaging intratumoral metabolic heterogeneity in specimens from patients with breast cancer. High-throughput single-cell photoacoustic microscopy of oxygen consumption rates should enable the faster characterization of intratumoral metabolic heterogeneity.
Figures

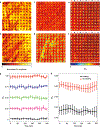
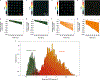
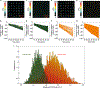
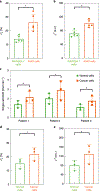

Comment in
-
Imaging intratumoral metabolic heterogeneity.Nat Biomed Eng. 2019 May;3(5):333-334. doi: 10.1038/s41551-019-0398-z. Nat Biomed Eng. 2019. PMID: 31073174 No abstract available.
References
-
- Almendro V, Marusyk A & Polyak K Cellular heterogeneity and molecular evolution in cancer. Annu. Rev. Pathol. 8, 277–302 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

