Identification of key genes, pathways, and miRNA/mRNA regulatory networks of CUMS-induced depression in nucleus accumbens by integrated bioinformatics analysis
- PMID: 30936699
- PMCID: PMC6421879
- DOI: 10.2147/NDT.S200264
Identification of key genes, pathways, and miRNA/mRNA regulatory networks of CUMS-induced depression in nucleus accumbens by integrated bioinformatics analysis
Abstract
Introduction: Major depressive disorder (MDD) is a recurrent, devastating mental disorder, which affects >350 million people worldwide, and exerts substantial public health and financial costs to society. Thus, there is a significant need to discover innovative therapeutics to treat depression efficiently. Stress-induced dysfunction in the subtype of neuronal cells and the change of synaptic plasticity and structural plasticity of nucleus accumbens (NAc) are implicated in depression symptomology. However, the molecular and epigenetic mechanisms and stresses to the NAc pathological changes in depression remain elusive.
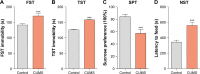
Materials and methods: In this study, treatment group mice were treated continually with the chronic unpredictable mild stress (CUMS) until expression of depression-like behaviors were found. Depression was confirmed with sucrose preference, novelty-suppressed feeding, forced swimming, and tail suspension tests. We applied high-throughput RNA sequencing to assess microRNA expression and transcriptional profiles in the NAc tissue from depression-like behaviors mice and control mice. The regulatory network of miRNAs/mRNAs was constructed based on the high-throughput RNA sequence and bioinformatics software predictions.
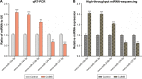
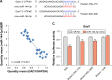
Results: A total of 17 miRNAs and 10 mRNAs were significantly upregulated in the NAc of CUMS-induced mice with depression-like behaviors, and 12 miRNAs and 29 mRNAs were downregulated. A series of bioinformatics analyses showed that these altered miRNAs predicted target mRNA and differentially expressed mRNAs were significantly enriched in the MAPK signaling pathway, GABAergic synapse, dopaminergic synapse, cytokine-cytokine receptor interaction, axon guidance, regulation of autophagy, and so on. Furthermore, dual luciferase report assay and qRT-PCR results validated the miRNA/mRNA regulatory network.
Conclusion: The deteriorations of GABAergic synapses, dopaminergic synapses, neurotransmitter synthesis, as well as autophagy-associated apoptotic pathway are associated with the molecular pathological mechanism of CUMS-induced depression.
Keywords: depression; mRNA; miRNA; nucleus accumbens; stress.
Conflict of interest statement
Disclosure The authors report no conflicts of interest in this work.
Figures

Similar articles
-
microRNA and mRNA profiles in nucleus accumbens underlying depression versus resilience in response to chronic stress.Am J Med Genet B Neuropsychiatr Genet. 2018 Sep;177(6):563-579. doi: 10.1002/ajmg.b.32651. Epub 2018 Aug 14. Am J Med Genet B Neuropsychiatr Genet. 2018. PMID: 30105773 Free PMC article.
-
The molecular mechanism underlying GABAergic dysfunction in nucleus accumbens of depression-like behaviours in mice.J Cell Mol Med. 2019 Oct;23(10):7021-7028. doi: 10.1111/jcmm.14596. Epub 2019 Aug 20. J Cell Mol Med. 2019. PMID: 31430030 Free PMC article.
-
Molecular Mechanism for Stress-Induced Depression Assessed by Sequencing miRNA and mRNA in Medial Prefrontal Cortex.PLoS One. 2016 Jul 18;11(7):e0159093. doi: 10.1371/journal.pone.0159093. eCollection 2016. PLoS One. 2016. PMID: 27427907 Free PMC article.
-
microRNA and mRNA profiles in the amygdala are associated with stress-induced depression and resilience in juvenile mice.Psychopharmacology (Berl). 2019 Jul;236(7):2119-2142. doi: 10.1007/s00213-019-05209-z. Epub 2019 Mar 21. Psychopharmacology (Berl). 2019. PMID: 30900007
-
Antidepressant-like effect of ginsenoside Rb1 on potentiating synaptic plasticity via the miR-134-mediated BDNF signaling pathway in a mouse model of chronic stress-induced depression.J Ginseng Res. 2022 May;46(3):376-386. doi: 10.1016/j.jgr.2021.03.005. Epub 2021 Mar 20. J Ginseng Res. 2022. PMID: 35600767 Free PMC article.
Cited by
-
Integrating weighted gene co-expression network analysis and machine learning to elucidate neural characteristics in a mouse model of depression.Front Psychiatry. 2025 Jun 27;16:1564095. doi: 10.3389/fpsyt.2025.1564095. eCollection 2025. Front Psychiatry. 2025. PMID: 40656047 Free PMC article.
-
The 24-Form Tai Chi Improves Anxiety and Depression and Upregulates miR-17-92 in Coronary Heart Disease Patients After Percutaneous Coronary Intervention.Front Physiol. 2020 Mar 11;11:149. doi: 10.3389/fphys.2020.00149. eCollection 2020. Front Physiol. 2020. PMID: 32218741 Free PMC article.
-
Deciphering the role of miRNA-134 in the pathophysiology of depression: A comprehensive review.Heliyon. 2024 Oct 5;10(19):e39026. doi: 10.1016/j.heliyon.2024.e39026. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39435111 Free PMC article. Review.
-
Life extension factor klotho regulates behavioral responses to stress via modulation of GluN2B function in the nucleus accumbens.Neuropsychopharmacology. 2022 Aug;47(9):1710-1720. doi: 10.1038/s41386-022-01323-3. Epub 2022 Apr 21. Neuropsychopharmacology. 2022. PMID: 35449449 Free PMC article.
-
Dynamic changes of behaviors, dentate gyrus neurogenesis and hippocampal miR-124 expression in rats with depression induced by chronic unpredictable mild stress.Neural Regen Res. 2020 Jun;15(6):1150-1159. doi: 10.4103/1673-5374.270414. Neural Regen Res. 2020. PMID: 31823896 Free PMC article.
References
-
- Anthes E. Depression: a change of mind. Nature. 2014;515(7526):185–187. - PubMed
-
- Smith K. Mental health: a world of depression. Nature. 2014;515(7526):180–181. - PubMed
-
- Gaynes BN, Warden D, Trivedi MH, Wisniewski SR, Fava M, Rush AJ. What did STAR*D teach us? Results from a large-scale, practical, clinical trial for patients with depression. Psychiatric Services. 2009;60(11):1439–1445. - PubMed
LinkOut - more resources
Full Text Sources

