Complete Genome Sequence of Halorubrum ezzemoulense Strain Fb21
- PMID: 30938699
- PMCID: PMC6430316
- DOI: 10.1128/MRA.00096-19
Complete Genome Sequence of Halorubrum ezzemoulense Strain Fb21
Abstract
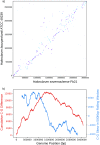
Isolated from Aran-Bidgol Lake in Iran, and reported here, Halorubrum ezzemoulense strain Fb21 represents the first complete genome from this archaeal species. Local recombination in this genome is in stark contrast to equidistant recombination events in bacteria. The genome's GC bias, however, points to a genome architecture and origin that resemble those of a bacterium. Its availability, genome signatures, and frequent intragenomic recombination mean that Fb21 presents an attractive model organism for this species.
Copyright © 2019 Feng et al.
Figures
References
-
- Corral P, de la Haba RR, Infante-Domínguez C, Sánchez-Porro C, Amoozegar MA, Papke RT, Ventosa A. 2018. Halorubrum chaoviator Mancinelli et al. 2009 is a later, heterotypic synonym of Halorubrum ezzemoulense Kharroub et al. 2006. Emended description of Halorubrum ezzemoulense Kharroub et al. 2006. Int J Syst Evol Microbiol 68:3657–3665. doi:10.1099/ijsem.0.003005. - DOI - PubMed
-
- de la Haba RR, Corral P, Sánchez-Porro C, Infante-Dominguez C, Makkay A, Ammozegar M, Ventosa A, Papke RT. 2018. Genotypic and lipid analyses of strains from the archaeal genus Halorubrum reveal insights into their taxonomy, divergence, and population structure. Front Microbiol 9:512. doi:10.3389/fmicb.2018.00512. - DOI - PMC - PubMed
-
- Dyall-Smith M. 2009. The Halohandbook: protocols for haloarchaeal genetics, v.7.2. Haloarchaeal Genetics Laboratory; https://haloarchaea.com/halohandbook/.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

