Genome-Wide Association Study of Susceptibility Loci for T-Cell Acute Lymphoblastic Leukemia in Children
- PMID: 30938820
- PMCID: PMC6910193
- DOI: 10.1093/jnci/djz043
Genome-Wide Association Study of Susceptibility Loci for T-Cell Acute Lymphoblastic Leukemia in Children
Abstract
Background: Acute lymphoblastic leukemia (ALL) is the most common cancer in children and can arise in B or T lymphoid lineages. Although risk loci have been identified for B-ALL, the inherited basis of T-ALL is mostly unknown, with a particular paucity of genome-wide investigation of susceptibility variants in large patient cohorts.
Methods: We performed a genome-wide association study (GWAS) in 1191 children with T-ALL and 12 178 controls, with independent replication using 117 cases and 5518 controls. The associations were tested using an additive logistic regression model. Top risk variants were tested for effects on enhancer activity using luciferase assay. All statistical tests were two sided.
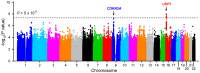
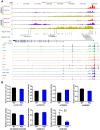
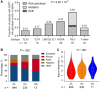
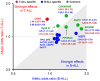
Results: A novel risk locus in the USP7 gene (rs74010351, odds ratio [OR] = 1.44, 95% confidence interval [CI] = 1.27 to 1.65, P = 4.51 × 10-8) reached genome-wide significance in the discovery cohort, with independent validation (OR = 1.51, 95% CI = 1.03 to 2.22, P = .04). The USP7 risk allele was overrepresented in individuals of African descent, thus contributing to the higher incidence of T-ALL in this race/ethnic group. Genetic changes in USP7 (germline variants or somatic mutations) were observed in 56.4% of T-ALL with TAL1 overexpression, statistically significantly higher than in any other subtypes. Functional analyses suggested this T-ALL risk allele is located in a putative cis-regulatory DNA element with negative effects on USP7 transcription. Finally, comprehensive comparison of 14 susceptibility loci in T- vs B-ALL pointed to distinctive etiology of these leukemias.
Conclusions: These findings indicate strong associations between inherited genetic variation and T-ALL susceptibility in children and shed new light on the molecular etiology of ALL, particularly commonalities and differences in the biology of the two major subtypes (B- vs T-ALL).
© The Author(s) 2019. Published by Oxford University Press. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures
References
-
- Linabery AM, Ross JA.. Trends in childhood cancer incidence in the U.S. (1992-2004). Cancer. 2008;112(2):416–432. - PubMed
-
- Hunger SP, Mullighan CG.. Acute lymphoblastic leukemia in children. N Engl J Med. 2015;373(16):1541–1552. - PubMed
-
- Pui CH, Sandlund JT, Pei D, et al. Results of therapy for acute lymphoblastic leukemia in black and white children. JAMA. 2003;290(15):2001–2007. - PubMed
-
- Pui CH, Behm FG, Singh B, et al. Heterogeneity of presenting features and their relation to treatment outcome in 120 children with T-cell acute lymphoblastic leukemia. Blood. 1990;75(1):174–179. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

