The role of the GATA transcription factor AreB in regulation of nitrogen and carbon metabolism in Aspergillus nidulans
- PMID: 30939206
- PMCID: PMC6494665
- DOI: 10.1093/femsle/fnz066
The role of the GATA transcription factor AreB in regulation of nitrogen and carbon metabolism in Aspergillus nidulans
Abstract
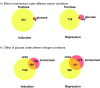
In Aspergillus nidulans, nitrogen and carbon metabolism are under the control of wide-domain regulatory systems, including nitrogen metabolite repression, carbon catabolite repression and the nutrient starvation response. Transcriptomic analysis of the wild type strain grown under different combinations of carbon and nitrogen regimes was performed, to identify differentially regulated genes. Carbon metabolism predominates as the most important regulatory signal but for many genes, both carbon and nitrogen metabolisms coordinate regulation. To identify mechanisms coordinating nitrogen and carbon metabolism, we tested the role of AreB, previously identified as a regulator of genes involved in nitrogen metabolism. Deletion of areB has significant phenotypic effects on the utilization of specific carbon sources, confirming its role in the regulation of carbon metabolism. AreB was shown to regulate the expression of areA, tamA, creA, xprG and cpcA regulatory genes suggesting areB has a range of indirect, regulatory effects. Different isoforms of AreB are produced as a result of differential splicing and use of two promoters which are differentially regulated by carbon and nitrogen conditions. These isoforms are likely to be functionally distinct and thus contributing to the modulation of AreB activity.
Keywords: AreA; AreB; CreA; Nitrogen metabolite repression; RNA-Seq; carbon catabolite repression.
© FEMS 2019.
Figures




References
-
- Abreu C, Sanguinetti M, Amillis Set al. .. UreA, the major urea/H+ symporter in Aspergillus nidulans. Fungal Genet Biol. 2010;7:1023–33. - PubMed
-
- Alam MA, Kelly JM. Proteins interacting with CreA and CreB in the carbon catabolite repression network in Aspergillus nidulans. Curr Genet. 2017;3:669–83. - PubMed
-
- Arst HN Jr., Cove DJ. Nitrogen metabolite repression in Aspergillus nidulans. Mol Gen Genet. 1973;126:111–41. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

