Germline deletion of ETV6 in familial acute lymphoblastic leukemia
- PMID: 30940639
- PMCID: PMC6457220
- DOI: 10.1182/bloodadvances.2018030635
Germline deletion of ETV6 in familial acute lymphoblastic leukemia
Abstract
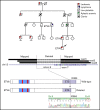
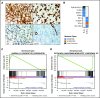
Recent studies have identified germline mutations in TP53, PAX5, ETV6, and IKZF1 in kindreds with familial acute lymphoblastic leukemia (ALL), but the genetic basis of ALL in many kindreds is unknown despite mutational analysis of the exome. Here, we report a germline deletion of ETV6 identified by linkage and structural variant analysis of whole-genome sequencing data segregating in a kindred with thrombocytopenia, B-progenitor acute lymphoblastic leukemia, and diffuse large B-cell lymphoma. The 75-nt deletion removed the ETV6 exon 7 splice acceptor, resulting in exon skipping and protein truncation. The ETV6 deletion was also identified by optimal structural variant analysis of exome sequencing data. These findings identify a new mechanism of germline predisposition in ALL and implicate ETV6 germline variation in predisposition to lymphoma. Importantly, these data highlight the importance of germline structural variant analysis in the search for germline variants predisposing to familial leukemia.
© 2019 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: C.G.M. has received consulting fees and travel funding from Amgen and Pfizer and research funding from AbbVie, Loxo Oncology, and Pfizer. The content of these activities and research is unrelated to the content of this manuscript. The remaining authors declare no competing financial interests.
Figures

References
-
- Mullighan CG, Goorha S, Radtke I, et al. . Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446(7137):758-764. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

