Yersinia pestis and plague: an updated view on evolution, virulence determinants, immune subversion, vaccination, and diagnostics
- PMID: 30940874
- PMCID: PMC6760536
- DOI: 10.1038/s41435-019-0065-0
Yersinia pestis and plague: an updated view on evolution, virulence determinants, immune subversion, vaccination, and diagnostics
Abstract
Plague is a vector-borne disease caused by Yersinia pestis. Transmitted by fleas from rodent reservoirs, Y. pestis emerged <6000 years ago from an enteric bacterial ancestor through events of gene gain and genome reduction. It is a highly remarkable model for the understanding of pathogenic bacteria evolution, and a major concern for public health as highlighted by recent human outbreaks. A complex set of virulence determinants, including the Yersinia outer-membrane proteins (Yops), the broad-range protease Pla, pathogen-associated molecular patterns (PAMPs), and iron capture systems play critical roles in the molecular strategies that Y. pestis employs to subvert the human immune system, allowing unrestricted bacterial replication in lymph nodes (bubonic plague) and in lungs (pneumonic plague). Some of these immunogenic proteins as well as the capsular antigen F1 are exploited for diagnostic purposes, which are critical in the context of the rapid onset of death in the absence of antibiotic treatment (less than a week for bubonic plague and <48 h for pneumonic plague). Here, we review recent research advances on Y. pestis evolution, virulence factor function, bacterial strategies to subvert mammalian innate immune responses, vaccination, and problems associated with pneumonic plague diagnosis.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

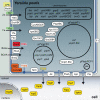
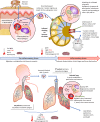
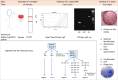
References
-
- Yersin A. La peste bubonique a Hong-Kong. Ann Inst Pasteur (Paris) 1894;8:662–7.
-
- Bertherat E. Plague around the world, 2010–2015. Wkly Epidemiol Rec. 2016;91:89–104.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

